4Q6F
 
 | | Crystal structure of human BAZ2A PHD zinc finger in complex with unmodified H3K4 histone peptide | | 分子名称: | 1,2-ETHANEDIOL, Bromodomain adjacent to zinc finger domain protein 2A, ZINC ION, ... | | 著者 | Tallant, C, Overvoorde, L, Krojer, T, Filippakopoulos, P, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Ciulli, A, Knapp, S, Structural Genomics Consortium (SGC) | | 登録日 | 2014-04-22 | | 公開日 | 2014-05-21 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.91 Å) | | 主引用文献 | Molecular basis of histone tail recognition by human TIP5 PHD finger and bromodomain of the chromatin remodeling complex NoRC.
Structure, 23, 2015
|
|
5ISB
 
 | | Crystal structure of mouse CARM1 in complex with inhibitor SA0435 | | 分子名称: | 1,2-ETHANEDIOL, 1-METHOXY-2-(2-METHOXYETHOXY)ETHANE, 5'-{[(3S)-3-amino-3-carboxypropyl][(1H-imidazol-4-yl)methyl]amino}-5'-deoxyadenosine, ... | | 著者 | Cura, V, Marechal, N, Mailliot, J, Troffer-Charlier, N, Hassenboehler, P, Wurtz, J.M, Bonnefond, L, Cavarelli, J. | | 登録日 | 2016-03-15 | | 公開日 | 2017-03-29 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystal structure of mouse CARM1 in complex with inhibitor SA0435
To Be Published
|
|
5IDK
 
 | |
4Q7O
 
 | | The crystal structure of an immunity protein NMB0503 from Neisseria meningitidis MC58 | | 分子名称: | BROMIDE ION, FORMIC ACID, Immunity protein | | 著者 | Tan, K, Stols, L, Eschenfeldt, W, Babnigg, G, Low, D.A, Hayes, C.S, Goulding, C.W, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Structure-Function Analysis of Polymorphic CDI Toxin-Immunity Protein Complexes (UC4CDI) | | 登録日 | 2014-04-25 | | 公開日 | 2014-05-14 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | The structure of a contact-dependent growth-inhibition (CDI) immunity protein from Neisseria meningitidis MC58.
Acta Crystallogr F Struct Biol Commun, 71, 2015
|
|
7STY
 
 | | Crystal structure of human CORO1C | | 分子名称: | Coronin-1C | | 著者 | Zeng, H, Dong, A, Hutchinson, A, Seitova, A, Loppnau, P, Arrowsmith, C.H, Edwards, A.M, Halabelian, L, Structural Genomics Consortium (SGC) | | 登録日 | 2021-11-15 | | 公開日 | 2021-12-01 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystal structure of human CORO1C
To Be Published
|
|
7SUL
 
 | | Crystal structure of the WD-repeat domain of human SEC31A | | 分子名称: | Protein transport protein Sec31A | | 著者 | Zeng, H, Dong, A, Loppnau, P, Hutchinson, A, Seitova, A, Arrowsmith, C.H, Edwards, A.M, Halabelian, L, Structural Genomics Consortium (SGC) | | 登録日 | 2021-11-17 | | 公開日 | 2021-12-01 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Crystal structure of the WD-repeat domain of human SEC31A
To Be Published
|
|
7T1Q
 
 | | Crystal Structure of the Succinyl-diaminopimelate Desuccinylase (DapE) from Acinetobacter baumannii in complex with Succinic Acid | | 分子名称: | ACETATE ION, SUCCINIC ACID, Succinyl-diaminopimelate desuccinylase, ... | | 著者 | Minasov, G, Shuvalova, L, Brunzelle, J.S, Dubrovska, I, Pshenychnyi, S, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2021-12-02 | | 公開日 | 2021-12-15 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Crystal Structure of the Succinyl-diaminopimelate Desuccinylase (DapE) from Acinetobacter baumannii in complex with Succinic Acid.
To be Published
|
|
4PW4
 
 | | Crystal structure of Aminopeptidase N in complex with phosphonic acid analogue of homophenylalanine L-(R)-hPheP | | 分子名称: | Aminopeptidase N, GLYCEROL, IMIDAZOLE, ... | | 著者 | Nocek, B, Mulligan, R, Vassiliou, S, Berlicki, L, Mucha, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2014-03-18 | | 公開日 | 2014-06-25 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Crystal structure of Aminopeptidase N in complex with phosphonic analogs of homophenylalanine
TO BE PUBLISHED
|
|
7T49
 
 | | Structure of SARS-CoV-2 3CL protease in complex with inhibitor 10c | | 分子名称: | (1R,2S)-1-hydroxy-2-{[N-({[7-(methanesulfonyl)-7-azaspiro[3.5]nonan-2-yl]oxy}carbonyl)-L-leucyl]amino}-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, (1S,2S)-1-hydroxy-2-{[N-({[7-(methanesulfonyl)-7-azaspiro[3.5]nonan-2-yl]oxy}carbonyl)-L-leucyl]amino}-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase | | 著者 | Lovell, S, Liu, L, Battaile, K.P, Chamandi, S.D, Kim, Y, Groutas, W.C, Chang, K.O. | | 登録日 | 2021-12-09 | | 公開日 | 2021-12-22 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Structure-Guided Design of Potent Spirocyclic Inhibitors of Severe Acute Respiratory Syndrome Coronavirus-2 3C-like Protease.
J.Med.Chem., 65, 2022
|
|
7SSE
 
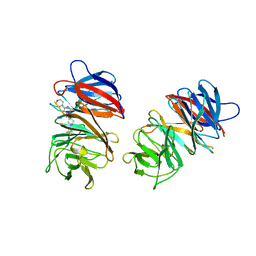 | | Crystal structure of the WDR domain of human DCAF1 in complex with CYCA-117-70 | | 分子名称: | DDB1- and CUL4-associated factor 1, N-[(3R)-1-(3-fluorophenyl)piperidin-3-yl]-6-(morpholin-4-yl)pyrimidin-4-amine | | 著者 | Kimani, S, Owen, J, Li, A, Dong, A, Li, Y, Hutchinson, A, Seitova, A, Shahani, V.M, Schapira, M, Arrowsmith, C.H, Edwards, A.M, Halabelian, L, Structural Genomics Consortium (SGC) | | 登録日 | 2021-11-10 | | 公開日 | 2021-12-15 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.62 Å) | | 主引用文献 | Discovery of a Novel DCAF1 Ligand Using a Drug-Target Interaction Prediction Model: Generalizing Machine Learning to New Drug Targets.
J.Chem.Inf.Model., 63, 2023
|
|
7T40
 
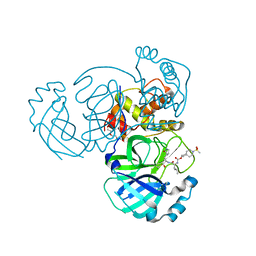 | | Structure of MERS 3CL protease in complex with inhibitor 10c | | 分子名称: | (1R,2S)-1-hydroxy-2-{[N-({[7-(methanesulfonyl)-7-azaspiro[3.5]nonan-2-yl]oxy}carbonyl)-L-leucyl]amino}-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, (1S,2S)-1-hydroxy-2-{[N-({[7-(methanesulfonyl)-7-azaspiro[3.5]nonan-2-yl]oxy}carbonyl)-L-leucyl]amino}-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase, ... | | 著者 | Lovell, S, Liu, L, Battaile, K.P, Chamandi, S.D, Kim, Y, Groutas, W.C, Chang, K.O. | | 登録日 | 2021-12-09 | | 公開日 | 2021-12-22 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structure-Guided Design of Potent Spirocyclic Inhibitors of Severe Acute Respiratory Syndrome Coronavirus-2 3C-like Protease.
J.Med.Chem., 65, 2022
|
|
7T42
 
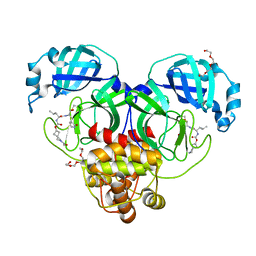 | | Structure of SARS-CoV-2 3CL protease in complex with inhibitor 2c | | 分子名称: | (1R,2S)-1-hydroxy-2-{[N-({[2-(2-methylpropanoyl)-2-azaspiro[3.3]heptan-6-yl]oxy}carbonyl)-L-leucyl]amino}-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, (1S,2S)-2-[(N-{[(2-acetyl-2-azaspiro[3.3]heptan-6-yl)oxy]carbonyl}-L-leucyl)amino]-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase, ... | | 著者 | Liu, L, Lovell, S, Battaile, K.P, Chamandi, S.D, Kim, Y, Groutas, W.C, Chang, K.O. | | 登録日 | 2021-12-09 | | 公開日 | 2021-12-22 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Structure-Guided Design of Potent Spirocyclic Inhibitors of Severe Acute Respiratory Syndrome Coronavirus-2 3C-like Protease.
J.Med.Chem., 65, 2022
|
|
4PXZ
 
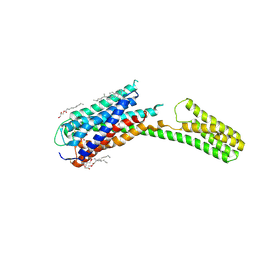 | | Crystal structure of P2Y12 receptor in complex with 2MeSADP | | 分子名称: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 2-(methylsulfanyl)adenosine 5'-(trihydrogen diphosphate), CHOLESTEROL, ... | | 著者 | Zhang, J, Zhang, K, Gao, Z.G, Paoletta, S, Zhang, D, Han, G.W, Li, T, Ma, L, Zhang, W, Muller, C.E, Yang, H, Jiang, H, Cherezov, V, Katritch, V, Jacobson, K.A, Stevens, R.C, Wu, B, Zhao, Q, GPCR Network (GPCR) | | 登録日 | 2014-03-25 | | 公開日 | 2014-04-30 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Agonist-bound structure of the human P2Y12 receptor
Nature, 509, 2014
|
|
5ICR
 
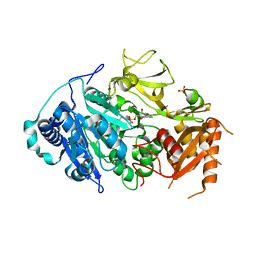 | | 2.25 Angstrom Resolution Crystal Structure of Fatty-Acid-CoA Ligase (FadD32) from Mycobacterium smegmatis in complex with Inhibitor 5'-O-[(11-phenoxyundecanoyl)sulfamoyl]adenosine. | | 分子名称: | 5'-O-[(11-phenoxyundecanoyl)sulfamoyl]adenosine, Acyl-CoA synthase, CHLORIDE ION, ... | | 著者 | Minasov, G, Shuvalova, L, Hung, D, Fisher, S.L, Edelstein, J, Kiryukhina, O, Dubrovska, I, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2016-02-23 | | 公開日 | 2016-04-06 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | 2.25 Angstrom Resolution Crystal Structure of Fatty-Acid-CoA Ligase (FadD32) from Mycobacterium smegmatis in complex with Inhibitor 5'-O-[(11-phenoxyundecanoyl)sulfamoyl]adenosine.
To Be Published
|
|
2LZZ
 
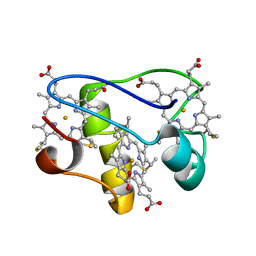 | | Solution structure of a mutant of the triheme cytochrome PpcA from Geobacter sulfurreducens sheds light on the role of the conserved aromatic residue F15 | | 分子名称: | Cytochrome c, 3 heme-binding sites, PROTOPORPHYRIN IX CONTAINING FE | | 著者 | Dantas, J.M, Morgado, L, Turner, D.L, Salgueiro, C.A. | | 登録日 | 2012-10-12 | | 公開日 | 2013-01-30 | | 最終更新日 | 2013-03-13 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of a mutant of the triheme cytochrome PpcA from Geobacter sulfurreducens sheds light on the role of the conserved aromatic residue F15.
Biochim.Biophys.Acta, 1827, 2013
|
|
7T39
 
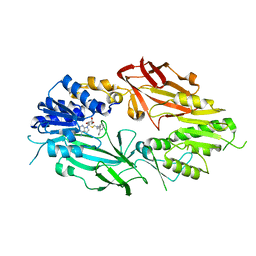 | | Co-crystal structure of human PRMT9 in complex with MT221 inhibitor | | 分子名称: | 7-[5-S-(4-{[(2-ethylpyridin-3-yl)methyl]amino}butyl)-5-thio-beta-D-ribofuranosyl]-7H-pyrrolo[2,3-d]pyrimidin-4-amine, Protein arginine N-methyltransferase 9 | | 著者 | Zeng, H, Dong, A, Hutchinson, A, Seitova, A, Li, Y, Gao, Y.D, Schneider, S, Siliphaivanh, P, Sloman, D, Nicholson, B, Fischer, C, Hicks, J, Brown, P.J, Arrowsmith, C.H, Edwards, A.M, Halabelian, L, Structural Genomics Consortium (SGC) | | 登録日 | 2021-12-07 | | 公開日 | 2021-12-22 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.81 Å) | | 主引用文献 | Co-crystal structure of human PRMT9 in complex with MT221 inhibitor
To Be Published
|
|
7T3Y
 
 | | Structure of MERS 3CL protease in complex with inhibitor 8c | | 分子名称: | (1R,2S)-1-hydroxy-2-{[N-({[7-(2-methylpropanoyl)-7-azaspiro[3.5]nonan-2-yl]oxy}carbonyl)-L-leucyl]amino}-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, (1S,2S)-1-hydroxy-2-{[N-({[7-(2-methylpropanoyl)-7-azaspiro[3.5]nonan-2-yl]oxy}carbonyl)-L-leucyl]amino}-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase | | 著者 | Lovell, S, Liu, L, Battaile, K.P, Chamandi, S.D, Kim, Y, Groutas, W.C, Chang, K.O. | | 登録日 | 2021-12-09 | | 公開日 | 2021-12-22 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structure-Guided Design of Potent Spirocyclic Inhibitors of Severe Acute Respiratory Syndrome Coronavirus-2 3C-like Protease.
J.Med.Chem., 65, 2022
|
|
1AJB
 
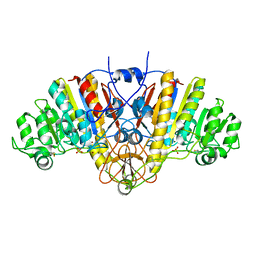 | | THREE-DIMENSIONAL STRUCTURE OF THE D153G MUTANT OF E. COLI ALKALINE PHOSPHATASE: A MUTANT WITH WEAKER MAGNESIUM BINDING AND INCREASED CATALYTIC ACTIVITY | | 分子名称: | ALKALINE PHOSPHATASE, MAGNESIUM ION, SULFATE ION, ... | | 著者 | Dealwis, C.G, Chen, L, Abad-Zapatero, C. | | 登録日 | 1995-08-19 | | 公開日 | 1995-11-14 | | 最終更新日 | 2021-11-03 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | 3-D structure of the D153G mutant of Escherichia coli alkaline phosphatase: an enzyme with weaker magnesium binding and increased catalytic activity.
Protein Eng., 8, 1995
|
|
7T4A
 
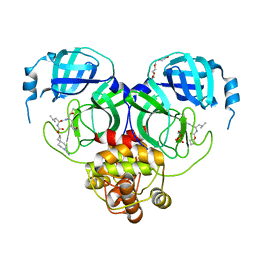 | | Structure of SARS-CoV-2 3CL protease in complex with inhibitor 11c | | 分子名称: | (1R,2S)-2-[(N-{[(7-cyano-7-azaspiro[3.5]nonan-2-yl)oxy]carbonyl}-L-leucyl)amino]-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, (1S,2S)-2-[(N-{[(7-cyano-7-azaspiro[3.5]nonan-2-yl)oxy]carbonyl}-L-leucyl)amino]-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase, ... | | 著者 | Lovell, S, Liu, L, Battaile, K.P, Chamandi, S.D, Kim, Y, Groutas, W.C, Chang, K.O. | | 登録日 | 2021-12-09 | | 公開日 | 2021-12-22 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structure-Guided Design of Potent Spirocyclic Inhibitors of Severe Acute Respiratory Syndrome Coronavirus-2 3C-like Protease.
J.Med.Chem., 65, 2022
|
|
7T46
 
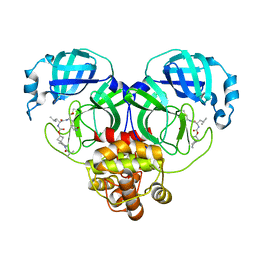 | | Structure of SARS-CoV-2 3CL protease in complex with inhibitor 8c | | 分子名称: | (1R,2S)-1-hydroxy-2-{[N-({[7-(2-methylpropanoyl)-7-azaspiro[3.5]nonan-2-yl]oxy}carbonyl)-L-leucyl]amino}-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, (1S,2S)-1-hydroxy-2-{[N-({[7-(2-methylpropanoyl)-7-azaspiro[3.5]nonan-2-yl]oxy}carbonyl)-L-leucyl]amino}-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase | | 著者 | Lovell, S, Liu, L, Battaile, K.P, Chamandi, S.D, Kim, Y, Groutas, W.C, Chang, K.O. | | 登録日 | 2021-12-09 | | 公開日 | 2021-12-22 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | Structure-Guided Design of Potent Spirocyclic Inhibitors of Severe Acute Respiratory Syndrome Coronavirus-2 3C-like Protease.
J.Med.Chem., 65, 2022
|
|
4XKJ
 
 | |
7T3Z
 
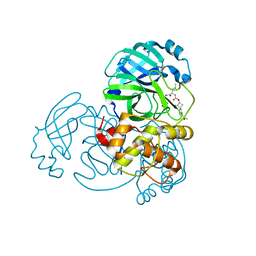 | | Structure of MERS 3CL protease in complex with inhibitor 9c | | 分子名称: | (1R,2S)-2-{[N-({[(2r,4R)-7-acetyl-7-azaspiro[3.5]non-5-en-2-yl]oxy}carbonyl)-L-leucyl]amino}-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, (1S,2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]-2-{[N-({[7-(phenylacetyl)-7-azaspiro[3.5]nonan-2-yl]oxy}carbonyl)-L-leucyl]amino}propane-1-sulfonic acid, 3C-like proteinase | | 著者 | Lovell, S, Liu, L, Battaile, K.P, Chamandi, S.D, Kim, Y, Groutas, W.C, Chang, K.O. | | 登録日 | 2021-12-09 | | 公開日 | 2021-12-22 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Structure-Guided Design of Potent Spirocyclic Inhibitors of Severe Acute Respiratory Syndrome Coronavirus-2 3C-like Protease.
J.Med.Chem., 65, 2022
|
|
7T48
 
 | | Structure of SARS-CoV-2 3CL protease in complex with inhibitor 9c | | 分子名称: | (1R,2S)-2-{[N-({[(2r,4R)-7-acetyl-7-azaspiro[3.5]non-5-en-2-yl]oxy}carbonyl)-L-leucyl]amino}-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, (1S,2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]-2-{[N-({[7-(phenylacetyl)-7-azaspiro[3.5]nonan-2-yl]oxy}carbonyl)-L-leucyl]amino}propane-1-sulfonic acid, 3C-like proteinase, ... | | 著者 | Lovell, S, Liu, L, Battaile, K.P, Chamandi, S.D, Kim, Y, Groutas, W.C, Chang, K.O. | | 登録日 | 2021-12-09 | | 公開日 | 2021-12-22 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structure-Guided Design of Potent Spirocyclic Inhibitors of Severe Acute Respiratory Syndrome Coronavirus-2 3C-like Protease.
J.Med.Chem., 65, 2022
|
|
7T43
 
 | | Structure of SARS-CoV-2 3CL protease in complex with inhibitor 3c | | 分子名称: | (1R,2S)-1-hydroxy-2-[(N-{[(2-methyl-2-azaspiro[3.3]heptan-6-yl)oxy]carbonyl}-L-leucyl)amino]-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, (1S,2S)-1-hydroxy-2-[(N-{[(2-methyl-2-azaspiro[3.3]heptan-6-yl)oxy]carbonyl}-L-leucyl)amino]-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase, ... | | 著者 | Liu, L, Lovell, S, Battaile, K.P, Chamandi, S.D, Kim, Y, Groutas, W.C, Chang, K.O. | | 登録日 | 2021-12-09 | | 公開日 | 2021-12-22 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structure-Guided Design of Potent Spirocyclic Inhibitors of Severe Acute Respiratory Syndrome Coronavirus-2 3C-like Protease.
J.Med.Chem., 65, 2022
|
|
7T88
 
 | | Crystal Structure of the C-terminal Domain of the Phosphate Acetyltransferase from Escherichia coli | | 分子名称: | 1,2-ETHANEDIOL, CHLORIDE ION, IODIDE ION, ... | | 著者 | Kim, Y, Dementiev, A, Welk, L, Endres, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2021-12-15 | | 公開日 | 2021-12-22 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Crystal Structure of c from Escherichia coli
To Be Published
|
|
