8YN7
 
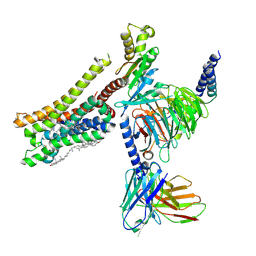 | | Cryo-EM structure of histamine H3 receptor in complex with immethridine and miniGo | | 分子名称: | 4-(1H-imidazol-4-ylmethyl)pyridine, Antibody fragment scFv16, CHOLESTEROL, ... | | 著者 | Zhang, X, Liu, G, Li, X, Gong, W. | | 登録日 | 2024-03-10 | | 公開日 | 2024-10-16 | | 最終更新日 | 2024-10-30 | | 実験手法 | ELECTRON MICROSCOPY (2.77 Å) | | 主引用文献 | Structural basis of ligand recognition and activation of the histamine receptor family.
Nat Commun, 15, 2024
|
|
8YN6
 
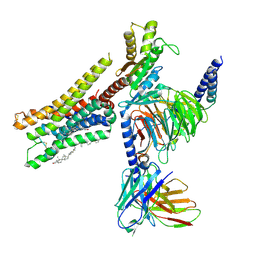 | | Cryo-EM structure of histamine H3 receptor in complex with imetit and Gi | | 分子名称: | 2-(1~{H}-imidazol-5-yl)ethyl carbamimidothioate, Antibody fragment scFv16, CHOLESTEROL, ... | | 著者 | Zhang, X, Liu, G, Li, X, Gong, W. | | 登録日 | 2024-03-10 | | 公開日 | 2024-10-09 | | 最終更新日 | 2024-11-27 | | 実験手法 | ELECTRON MICROSCOPY (2.77 Å) | | 主引用文献 | Structural basis of ligand recognition and activation of the histamine receptor family.
Nat Commun, 15, 2024
|
|
8YN2
 
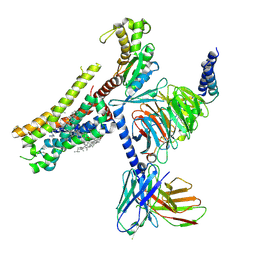 | | Cryo-EM structure of histamine H1 receptor in complex with histamine and miniGq | | 分子名称: | Antibody fragment scFv16, CHOLESTEROL, Engineered guanine nucleotide-binding protein G(q) subunit alpha, ... | | 著者 | Zhang, X, Liu, G, Li, X, Gong, W. | | 登録日 | 2024-03-10 | | 公開日 | 2024-10-09 | | 最終更新日 | 2024-11-20 | | 実験手法 | ELECTRON MICROSCOPY (2.66 Å) | | 主引用文献 | Structural basis of ligand recognition and activation of the histamine receptor family.
Nat Commun, 15, 2024
|
|
8YN9
 
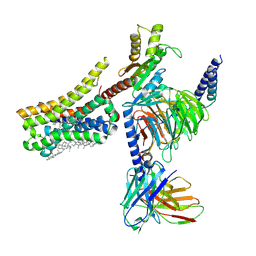 | | Cryo-EM structure of histamine H4 receptor in complex with histamine and Gi | | 分子名称: | Antibody fragment scFv16, CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | 著者 | Zhang, X, Liu, G, Li, X, Gong, W. | | 登録日 | 2024-03-10 | | 公開日 | 2024-10-09 | | 最終更新日 | 2024-11-06 | | 実験手法 | ELECTRON MICROSCOPY (2.3 Å) | | 主引用文献 | Structural basis of ligand recognition and activation of the histamine receptor family.
Nat Commun, 15, 2024
|
|
8YN5
 
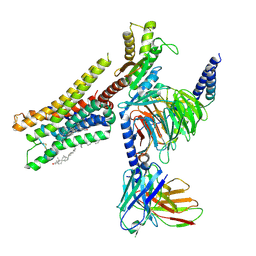 | | Cryo-EM structure of histamine H3 receptor in complex with histamine and Gi | | 分子名称: | Antibody fragment scFv16, CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | 著者 | Zhang, X, Liu, G, Li, X, Gong, W. | | 登録日 | 2024-03-10 | | 公開日 | 2024-10-09 | | 最終更新日 | 2024-11-27 | | 実験手法 | ELECTRON MICROSCOPY (2.7 Å) | | 主引用文献 | Structural basis of ligand recognition and activation of the histamine receptor family.
Nat Commun, 15, 2024
|
|
8YN3
 
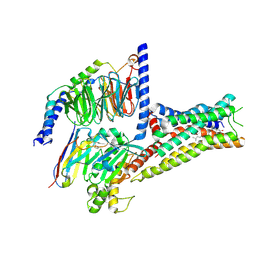 | | Cryo-EM structure of histamine H2 receptor in complex with histamine and miniGs | | 分子名称: | CHOLESTEROL, Engineered guanine nucleotide,binding protein G(s) subunit alpha, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | 著者 | Zhang, X, Liu, G, Li, X, Gong, W. | | 登録日 | 2024-03-10 | | 公開日 | 2024-10-09 | | 最終更新日 | 2024-11-06 | | 実験手法 | ELECTRON MICROSCOPY (2.56 Å) | | 主引用文献 | Structural basis of ligand recognition and activation of the histamine receptor family.
Nat Commun, 15, 2024
|
|
8YNA
 
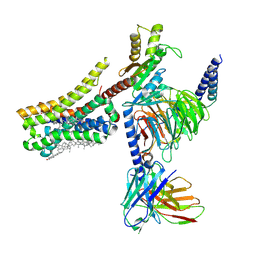 | | Cryo-EM structure of histamine H4 receptor in complex with immepip and Gi | | 分子名称: | 4-(1H-imidazol-5-ylmethyl)piperidine, Antibody fragment scFv16, CHOLESTEROL, ... | | 著者 | Zhang, X, Liu, G, Li, X, Gong, W. | | 登録日 | 2024-03-10 | | 公開日 | 2024-10-09 | | 最終更新日 | 2024-11-20 | | 実験手法 | ELECTRON MICROSCOPY (2.63 Å) | | 主引用文献 | Structural basis of ligand recognition and activation of the histamine receptor family.
Nat Commun, 15, 2024
|
|
8IZB
 
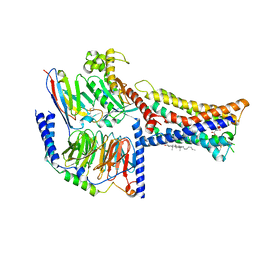 | | Lysophosphatidylserine receptor GPR174-Gs complex | | 分子名称: | CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | 著者 | Gong, W, Liu, G, Li, X, Zhang, X. | | 登録日 | 2023-04-06 | | 公開日 | 2023-11-01 | | 最終更新日 | 2024-10-30 | | 実験手法 | ELECTRON MICROSCOPY (3.06 Å) | | 主引用文献 | Structural basis for ligand recognition and signaling of the lysophosphatidylserine receptors GPR34 and GPR174.
Plos Biol., 21, 2023
|
|
8IZ4
 
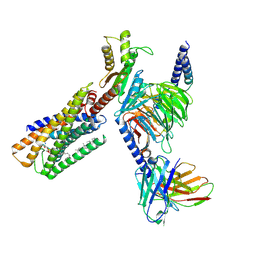 | | Lysophosphatidylserine receptor GPR34-Gi complex | | 分子名称: | Antibody fragment scFv16, CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | 著者 | Gong, W, Liu, G, Li, X, Zhang, X. | | 登録日 | 2023-04-06 | | 公開日 | 2024-04-10 | | 最終更新日 | 2024-10-30 | | 実験手法 | ELECTRON MICROSCOPY (2.93 Å) | | 主引用文献 | Structural mechanisms of ligand binding and signaling in lysophosphatidylserine receptors
To Be Published
|
|
8JCB
 
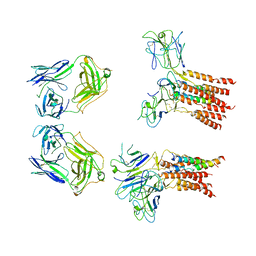 | | Vgamma5 Vdelta1 T cell receptor complex | | 分子名称: | T cell receptor delta variable 1,T cell receptor delta constant, T cell receptor gamma variable 5,T cell receptor gamma constant 1, T-cell surface glycoprotein CD3 delta chain, ... | | 著者 | Xin, W, Huang, B, Chi, X, Xu, M, Zhang, Y, Li, X, Su, Q, Zhou, Q. | | 登録日 | 2023-05-10 | | 公開日 | 2024-05-08 | | 最終更新日 | 2024-11-06 | | 実験手法 | ELECTRON MICROSCOPY (9.5 Å) | | 主引用文献 | Structures of human gamma delta T cell receptor-CD3 complex.
Nature, 630, 2024
|
|
8JC0
 
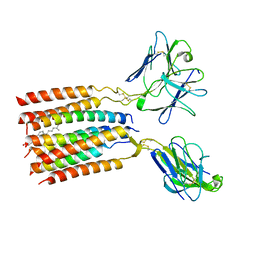 | | V gamma9 V delta2 TCR and CD3 complex in LMNG | | 分子名称: | CHOLESTEROL, T cell receptor delta variable 2,T cell receptor delta constant, T cell receptor gamma variable 9,T cell receptor gamma constant 1, ... | | 著者 | Xin, W, Huang, B, Chi, X, Xu, M, Zhang, Y, Li, X, Su, Q, Zhou, Q. | | 登録日 | 2023-05-10 | | 公開日 | 2024-05-08 | | 最終更新日 | 2024-11-06 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Structures of human gamma delta T cell receptor-CD3 complex.
Nature, 630, 2024
|
|
8JWE
 
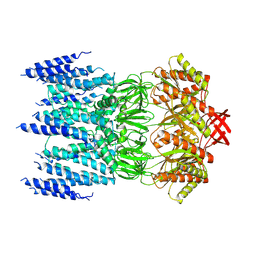 | |
8J00
 
 | | Human KCNQ2-CaM in complex with CBD | | 分子名称: | Calmodulin-1, Potassium voltage-gated channel subfamily KQT member 2, cannabidiol | | 著者 | Ma, D, Li, X, Guo, J. | | 登録日 | 2023-04-09 | | 公開日 | 2023-11-29 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Ligand activation mechanisms of human KCNQ2 channel.
Nat Commun, 14, 2023
|
|
8IZY
 
 | | Human KCNQ2-CaM in complex with HN37 | | 分子名称: | Potassium voltage-gated channel subfamily KQT member 2, methyl N-[4-[(4-fluorophenyl)methyl-prop-2-ynyl-amino]-2,6-dimethyl-phenyl]carbamate | | 著者 | Ma, D, Li, X, Guo, J. | | 登録日 | 2023-04-09 | | 公開日 | 2023-11-29 | | 実験手法 | ELECTRON MICROSCOPY (2.5 Å) | | 主引用文献 | Ligand activation mechanisms of human KCNQ2 channel.
Nat Commun, 14, 2023
|
|
8J04
 
 | | Human KCNQ2-CaM-HN37 complex in the presence of PIP2 | | 分子名称: | Calmodulin-1, Potassium voltage-gated channel subfamily KQT member 2, methyl N-[4-[(4-fluorophenyl)methyl-prop-2-ynyl-amino]-2,6-dimethyl-phenyl]carbamate | | 著者 | Ma, D, Li, X, Guo, J. | | 登録日 | 2023-04-09 | | 公開日 | 2023-12-06 | | 実験手法 | ELECTRON MICROSCOPY (2.7 Å) | | 主引用文献 | Ligand activation mechanisms of human KCNQ2 channel.
Nat Commun, 14, 2023
|
|
8J05
 
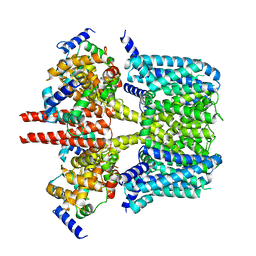 | |
8J02
 
 | | Human KCNQ2(F104A)-CaM-PIP2-CBD complex in state II | | 分子名称: | Calmodulin-1, Potassium voltage-gated channel subfamily KQT member 2, [(2R)-2-octanoyloxy-3-[oxidanyl-[(1R,2R,3S,4R,5R,6S)-2,3,6-tris(oxidanyl)-4,5-diphosphonooxy-cyclohexyl]oxy-phosphoryl]oxy-propyl] octanoate, ... | | 著者 | Ma, D, Li, X, Guo, J. | | 登録日 | 2023-04-09 | | 公開日 | 2023-12-13 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | Ligand activation mechanisms of human KCNQ2 channel.
Nat Commun, 14, 2023
|
|
8J01
 
 | | Human KCNQ2-CaM in complex with CBD and PIP2 | | 分子名称: | Calmodulin-1, Potassium voltage-gated channel subfamily KQT member 2, [(2R)-2-octanoyloxy-3-[oxidanyl-[(1R,2R,3S,4R,5R,6S)-2,3,6-tris(oxidanyl)-4,5-diphosphonooxy-cyclohexyl]oxy-phosphoryl]oxy-propyl] octanoate, ... | | 著者 | Ma, D, Li, X, Guo, J. | | 登録日 | 2023-04-09 | | 公開日 | 2023-12-13 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Ligand activation mechanisms of human KCNQ2 channel.
Nat Commun, 14, 2023
|
|
7BW7
 
 | | Cryo-EM Structure for the Ectodomain of the Full-length Human Insulin Receptor in Complex with 1 Insulin. | | 分子名称: | Insulin fusion, Insulin receptor | | 著者 | Yu, D, Zhang, X, Sun, J, Li, X, Wu, Z, Han, X, Fan, C, Ma, Y, Ouyang, Q, Wang, T. | | 登録日 | 2020-04-13 | | 公開日 | 2021-04-14 | | 最終更新日 | 2024-11-13 | | 実験手法 | ELECTRON MICROSCOPY (4.1 Å) | | 主引用文献 | Insulin Binding Induced the Ectodomain Conformational Dynamics in the Full-length Human Insulin Receptor
To Be Published
|
|
7BW8
 
 | | Cryo-EM Structure for the Insulin Binding Region in the Ectodomain of the Full-length Human Insulin Receptor in Complex with 1 Insulin | | 分子名称: | Insulin fusion, Insulin receptor | | 著者 | Yu, D, Zhang, X, Sun, J, Li, X, Wu, Z, Han, X, Fan, C, Ma, Y, Ouyang, Q, Wang, T. | | 登録日 | 2020-04-14 | | 公開日 | 2021-04-14 | | 最終更新日 | 2024-10-23 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Insulin Binding Induced the Ectodomain Conformational Dynamics in the Full-length Human Insulin Receptor
To Be Published
|
|
7BWA
 
 | | Cryo-EM Structure for the Ectodomain of the Full-length Human Insulin Receptor in Complex with 2 Insulin | | 分子名称: | Insulin fusion, Insulin receptor | | 著者 | Yu, D, Zhang, X, Sun, J, Li, X, Wu, Z, Han, X, Fan, C, Ma, Y, Ouyang, Q, Wang, T. | | 登録日 | 2020-04-14 | | 公開日 | 2021-04-14 | | 最終更新日 | 2024-11-06 | | 実験手法 | ELECTRON MICROSCOPY (4.9 Å) | | 主引用文献 | Insulin Binding Induced the Ectodomain Conformational Dynamics in the Full-length Human Insulin Receptor
To Be Published
|
|
8WZ2
 
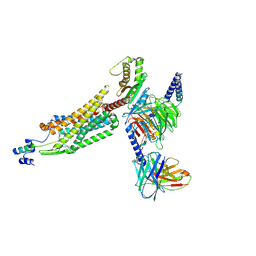 | | Structure of 26RFa-pyroglutamylated RFamide peptide receptor complex | | 分子名称: | G-alpha q, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | 著者 | Jin, S, Li, X, Xu, Y, Guo, S, Wu, C, Zhang, H, Yuan, Q, Xu, H.E, Xie, X, Jiang, Y. | | 登録日 | 2023-11-01 | | 公開日 | 2024-06-19 | | 最終更新日 | 2024-11-20 | | 実験手法 | ELECTRON MICROSCOPY (2.73 Å) | | 主引用文献 | Structural basis for recognition of 26RFa by the pyroglutamylated RFamide peptide receptor.
Cell Discov, 10, 2024
|
|
8WJ3
 
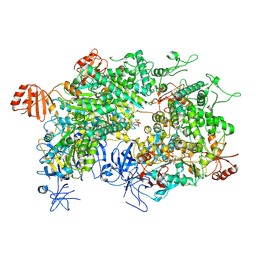 | | Cryo-EM structure of a bacterial protein | | 分子名称: | Helicase HerA central domain-containing protein, MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER | | 著者 | Yu, G, Liao, F, Li, X, Li, Z, Zhang, H. | | 登録日 | 2023-09-25 | | 公開日 | 2024-09-11 | | 最終更新日 | 2025-07-02 | | 実験手法 | ELECTRON MICROSCOPY (3.36 Å) | | 主引用文献 | Cryo-EM structure of a bacterial protein
To Be Published
|
|
8WLD
 
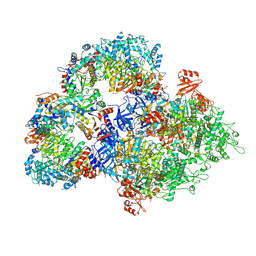 | | Cryo-EM structure of SIR2/HerA antiphage complex | | 分子名称: | Helicase HerA central domain-containing protein, SIR2-like domain-containing protein | | 著者 | Yu, G, Liao, F, Li, X, Li, Z, Zhang, H. | | 登録日 | 2023-09-29 | | 公開日 | 2024-09-11 | | 最終更新日 | 2025-07-02 | | 実験手法 | ELECTRON MICROSCOPY (3.48 Å) | | 主引用文献 | Cryo-EM structure of SIR2/HerA antiphage complex
To Be Published
|
|
8WOC
 
 | | Cryo-EM structure of SIR2/HerA complex | | 分子名称: | Helicase HerA central domain-containing protein, SIR2-like domain-containing protein | | 著者 | Yu, G, Liao, F, Li, X, Li, Z, Zhang, H. | | 登録日 | 2023-10-07 | | 公開日 | 2024-09-11 | | 最終更新日 | 2025-07-02 | | 実験手法 | ELECTRON MICROSCOPY (3.28 Å) | | 主引用文献 | Cryo-EM structure of a bacterial protein
To Be Published
|
|
