6IVU
 
 | |
6IVS
 
 | |
7EZP
 
 | | Indole-2-carboxylic acid derivatives as allosteric inhibitors of fructose-1,6-bisphosphatase | | 分子名称: | 1,6-di-O-phosphono-beta-D-fructofuranose, 3-(3-hydroxy-3-oxopropyl)-5-(2-methylpropyl)-7-nitro-1H-indole-2-carboxylic acid, Fructose-1,6-bisphosphatase 1 | | 著者 | Wang, X.Y, Zhou, J, Xu, B.L. | | 登録日 | 2021-06-01 | | 公開日 | 2022-06-01 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Discovery of Novel Indole Derivatives as Fructose-1,6-bisphosphatase Inhibitors and X-ray Cocrystal Structures Analysis.
Acs Med.Chem.Lett., 13, 2022
|
|
7EZF
 
 | | Indole-2-carboxylic acid derivatives as allosteric inhibitors of fructose-1,6-bisphosphatase | | 分子名称: | 1,6-di-O-phosphono-beta-D-fructofuranose, 7-chloranyl-5-ethyl-3-(3-hydroxy-3-oxopropyl)-1H-indole-2-carboxylic acid, Fructose-1,6-bisphosphatase 1 | | 著者 | Wang, X.Y, Zhou, J, Xu, B.L. | | 登録日 | 2021-06-01 | | 公開日 | 2022-06-01 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.76 Å) | | 主引用文献 | Discovery of Novel Indole Derivatives as Fructose-1,6-bisphosphatase Inhibitors and X-ray Cocrystal Structures Analysis.
Acs Med.Chem.Lett., 13, 2022
|
|
7EZR
 
 | | Indole-2-carboxylic acid derivatives as allosteric inhibitors of fructose-1,6-bisphosphatase | | 分子名称: | 1,6-di-O-phosphono-beta-D-fructofuranose, 5-ethyl-7-nitro-3-[3-oxidanylidene-3-(thiophen-2-ylsulfonylamino)propyl]-1H-indole-2-carboxylic acid, Fructose-1,6-bisphosphatase 1 | | 著者 | Wang, X.Y, Zhou, J, Xu, B.L. | | 登録日 | 2021-06-01 | | 公開日 | 2022-06-01 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (3.27 Å) | | 主引用文献 | Discovery of Novel Indole Derivatives as Fructose-1,6-bisphosphatase Inhibitors and X-ray Cocrystal Structures Analysis.
Acs Med.Chem.Lett., 13, 2022
|
|
7V69
 
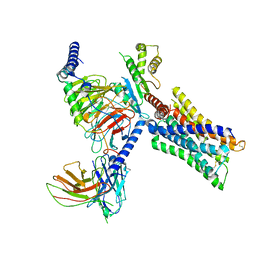 | | Cryo-EM structure of a class A GPCR-G protein complex | | 分子名称: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | 著者 | Wang, J.J, Wu, M, Wu, L.J, Hua, T, Liu, Z.J, Wang, T. | | 登録日 | 2021-08-20 | | 公開日 | 2022-05-11 | | 最終更新日 | 2022-11-23 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | The unconventional activation of the muscarinic acetylcholine receptor M4R by diverse ligands.
Nat Commun, 13, 2022
|
|
7V6A
 
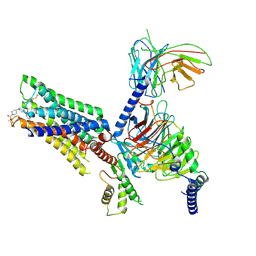 | | Cry-EM structure of M4-c110-G protein complex | | 分子名称: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | 著者 | Wang, J.J, Wu, M, Wu, L.J, Hua, T, Liu, Z.J, Wang, T. | | 登録日 | 2021-08-20 | | 公開日 | 2022-05-11 | | 最終更新日 | 2022-11-23 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | The unconventional activation of the muscarinic acetylcholine receptor M4R by diverse ligands.
Nat Commun, 13, 2022
|
|
7V68
 
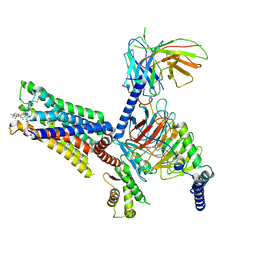 | | An Agonist and PAM-bound Class A GPCR with Gi protein complex structure | | 分子名称: | 3-amino-5-chloro-N-cyclopropyl-4-methyl-6-[2-(4-methylpiperazin-1-yl)-2-oxoethoxy]thieno[2,3-b]pyridine-2-carboxamide, 4-(4,5-dihydro-1,2-oxazol-3-yloxy)-N,N,N-trimethylbut-2-yn-1-aminium, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | 著者 | Wang, J.J, Wu, L.J, Wu, M, Hua, T, Liu, Z.J, Wang, T. | | 登録日 | 2021-08-20 | | 公開日 | 2022-05-11 | | 最終更新日 | 2022-11-23 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | The unconventional activation of the muscarinic acetylcholine receptor M4R by diverse ligands.
Nat Commun, 13, 2022
|
|
6KUR
 
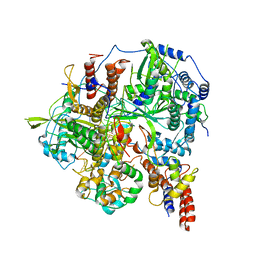 | | Structure of influenza D virus polymerase bound to vRNA promoter in Mode B conformation (Class B1) | | 分子名称: | 3'-vRNA, 5'-vRNA, Polymerase 3, ... | | 著者 | Peng, Q, Peng, R, Qi, J, Gao, G.F, Shi, Y. | | 登録日 | 2019-09-02 | | 公開日 | 2019-10-02 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Structural insight into RNA synthesis by influenza D polymerase.
Nat Microbiol, 4, 2019
|
|
6KUK
 
 | | Structure of influenza D virus polymerase bound to vRNA promoter in mode A conformation (class A1) | | 分子名称: | 3'-vRNA, 5'-vRNA, Polymerase 3, ... | | 著者 | Peng, Q, Peng, R, Qi, J, Gao, G.F, Shi, Y. | | 登録日 | 2019-09-02 | | 公開日 | 2019-10-02 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | Structural insight into RNA synthesis by influenza D polymerase.
Nat Microbiol, 4, 2019
|
|
6KUP
 
 | | Structure of influenza D virus polymerase bound to vRNA promoter in Mode A conformation(Class A2) | | 分子名称: | 3'-vRNA, 5'-vRNA, Polymerase 3, ... | | 著者 | Peng, Q, Peng, R, Qi, J, Gao, G.F, Shi, Y. | | 登録日 | 2019-09-02 | | 公開日 | 2019-10-02 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (4.3 Å) | | 主引用文献 | Structural insight into RNA synthesis by influenza D polymerase.
Nat Microbiol, 4, 2019
|
|
6KUV
 
 | | Structure of influenza D virus polymerase bound to cRNA promoter in class 2 | | 分子名称: | 3'-cRNA, 5'-cRNA, Polymerase 3, ... | | 著者 | Peng, Q, Peng, R, Qi, J, Gao, G.F, Shi, Y. | | 登録日 | 2019-09-02 | | 公開日 | 2019-10-02 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (4.1 Å) | | 主引用文献 | Structural insight into RNA synthesis by influenza D polymerase.
Nat Microbiol, 4, 2019
|
|
6KUT
 
 | | Structure of influenza D virus polymerase bound to vRNA promoter in Mode B conformation (Class B2) | | 分子名称: | 3'-vRNA, 5'-vRNA, Polymerase 3, ... | | 著者 | Peng, Q, Peng, R, Qi, J, Gao, G.F, Shi, Y. | | 登録日 | 2019-09-02 | | 公開日 | 2019-10-02 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (4.1 Å) | | 主引用文献 | Structural insight into RNA synthesis by influenza D polymerase.
Nat Microbiol, 4, 2019
|
|
6KV5
 
 | | Structure of influenza D virus apo polymerase | | 分子名称: | Polymerase 3, Polymerase PB2, RNA-directed RNA polymerase catalytic subunit | | 著者 | Peng, Q, Peng, R, Qi, J, Gao, G.F, Shi, Y. | | 登録日 | 2019-09-03 | | 公開日 | 2019-10-02 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (4.6 Å) | | 主引用文献 | Structural insight into RNA synthesis by influenza D polymerase.
Nat Microbiol, 4, 2019
|
|
2MG9
 
 | | Truncated EGF-A | | 分子名称: | CALCIUM ION, Low-density lipoprotein receptor | | 著者 | Schroeder, C.I, Rosengren, K. | | 登録日 | 2013-10-30 | | 公開日 | 2014-04-02 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Design and Synthesis of Truncated EGF-A Peptides that Restore LDL-R Recycling in the Presence of PCSK9 In Vitro.
Chem.Biol., 21, 2014
|
|
7F8L
 
 | |
7F5F
 
 | | SARS-CoV-2 ORF8 S84 | | 分子名称: | CALCIUM ION, ORF8 protein | | 著者 | Chen, S, Zhou, Z, Chen, X. | | 登録日 | 2021-06-22 | | 公開日 | 2022-01-19 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.62 Å) | | 主引用文献 | Crystal Structures of Bat and Human Coronavirus ORF8 Protein Ig-Like Domain Provide Insights Into the Diversity of Immune Responses.
Front Immunol, 12, 2021
|
|
4TQ6
 
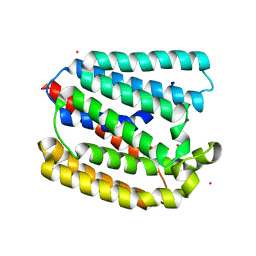 | | Structure of a UbiA homolog from Archaeoglobus fulgidus bound to Cd2+ | | 分子名称: | CADMIUM ION, prenyltransferase | | 著者 | Huang, H, Levin, E.J, Bai, Y, Zhou, M, New York Consortium on Membrane Protein Structure (NYCOMPS) | | 登録日 | 2014-06-10 | | 公開日 | 2014-07-16 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (3.0678 Å) | | 主引用文献 | Structure of a Membrane-Embedded Prenyltransferase Homologous to UBIAD1.
Plos Biol., 12, 2014
|
|
4TQ5
 
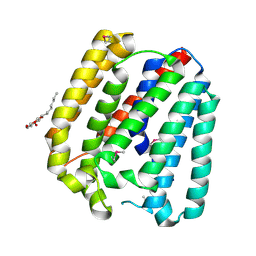 | | Structure of a UbiA homolog from Archaeoglobus fulgidus | | 分子名称: | octyl beta-D-glucopyranoside, prenyltransferase | | 著者 | Huang, H, Levin, E.J, Bai, Y, Zhou, M, New York Consortium on Membrane Protein Structure (NYCOMPS) | | 登録日 | 2014-06-10 | | 公開日 | 2014-07-16 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (3.2023 Å) | | 主引用文献 | Structure of a Membrane-Embedded Prenyltransferase Homologous to UBIAD1.
Plos Biol., 12, 2014
|
|
4TQ4
 
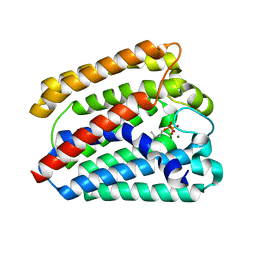 | | Structure of a UbiA homolog from Archaeoglobus fulgidus bound to DMAPP and Mg2+ | | 分子名称: | DIMETHYLALLYL DIPHOSPHATE, MAGNESIUM ION, prenyltransferase | | 著者 | Huang, H, Levin, E.J, Bai, Y, Zhou, M, New York Consortium on Membrane Protein Structure (NYCOMPS) | | 登録日 | 2014-06-10 | | 公開日 | 2014-07-16 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.5025 Å) | | 主引用文献 | Structure of a Membrane-Embedded Prenyltransferase Homologous to UBIAD1.
Plos Biol., 12, 2014
|
|
4TQ3
 
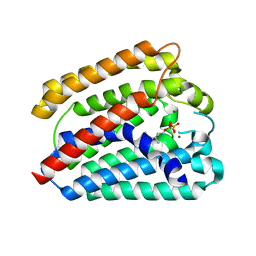 | | Structure of a UbiA homolog from Archaeoglobus fulgidus bound to GPP and Mg2+ | | 分子名称: | GERANYL DIPHOSPHATE, MAGNESIUM ION, Prenyltransferase | | 著者 | Huang, H, Levin, E.J, Bai, Y, Zhou, M, New York Consortium on Membrane Protein Structure (NYCOMPS) | | 登録日 | 2014-06-10 | | 公開日 | 2014-07-16 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.4076 Å) | | 主引用文献 | Structure of a Membrane-Embedded Prenyltransferase Homologous to UBIAD1.
Plos Biol., 12, 2014
|
|
8OYU
 
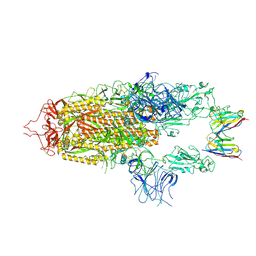 | | Stabilised BA.1 SARS-CoV-2 spike with H6 nanobodies in '2 up 1 down' RBD conformation | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, H6 nanobody, ... | | 著者 | Weckener, M, Naismith, J.H, Owens, R.J. | | 登録日 | 2023-05-05 | | 公開日 | 2024-05-15 | | 最終更新日 | 2024-06-26 | | 実験手法 | ELECTRON MICROSCOPY (4 Å) | | 主引用文献 | Structural and functional characterization of nanobodies that neutralize Omicron variants of SARS-CoV-2.
Open Biology, 14, 2024
|
|
8OWT
 
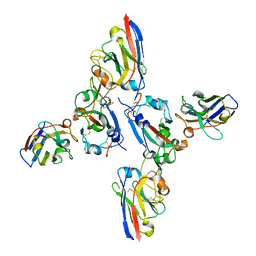 | | SARS-CoV-2 spike RBD with A8 and H3 nanobodies bound | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, Nanobody A8, ... | | 著者 | Mikolajek, H, Naismith, J.H, Owens, R.J. | | 登録日 | 2023-04-28 | | 公開日 | 2024-05-08 | | 最終更新日 | 2024-06-19 | | 実験手法 | X-RAY DIFFRACTION (2.37 Å) | | 主引用文献 | Structural and functional characterization of nanobodies that neutralize Omicron variants of SARS-CoV-2.
Open Biology, 14, 2024
|
|
8OWW
 
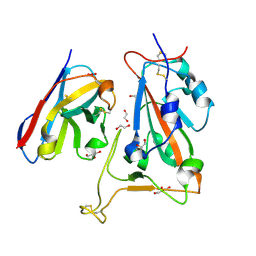 | | B5-5 nanobody bound to SARS-CoV-2 spike RBD (Wuhan) | | 分子名称: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, B5-5 nanobody, ... | | 著者 | Cornish, K.A.S, Naismith, J.H, Owens, R.J. | | 登録日 | 2023-04-28 | | 公開日 | 2024-05-08 | | 最終更新日 | 2024-06-19 | | 実験手法 | X-RAY DIFFRACTION (1.969 Å) | | 主引用文献 | Structural and functional characterization of nanobodies that neutralize Omicron variants of SARS-CoV-2.
Open Biology, 14, 2024
|
|
8OWV
 
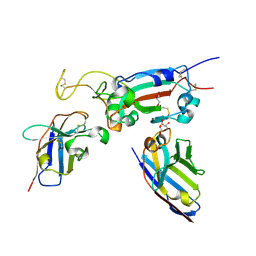 | | H6 and F2 nanobodies bound to SARS-CoV-2 spike RBD | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, F2, GLYCEROL, ... | | 著者 | Mikolajek, H, Naismith, J.H, Owens, R.J. | | 登録日 | 2023-04-28 | | 公開日 | 2024-05-08 | | 最終更新日 | 2024-06-19 | | 実験手法 | X-RAY DIFFRACTION (1.73 Å) | | 主引用文献 | Structural and functional characterization of nanobodies that neutralize Omicron variants of SARS-CoV-2.
Open Biology, 14, 2024
|
|
