3T6O
 
 | | The Structure of an Anti-sigma-factor antagonist (STAS) domain protein from Planctomyces limnophilus. | | 分子名称: | CHLORIDE ION, Sulfate transporter/antisigma-factor antagonist STAS | | 著者 | Cuff, M.E, Moser, C, Hatzos-Skintges, C, Bearden, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2011-07-28 | | 公開日 | 2011-09-07 | | 最終更新日 | 2017-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | The Structure of an Anti-sigma-factor antagonist (STAS) domain protein from Planctomyces limnophilus.
TO BE PUBLISHED
|
|
3SQN
 
 | | Putative Mga family transcriptional regulator from Enterococcus faecalis | | 分子名称: | Conserved domain protein | | 著者 | Osipiuk, J, Wu, R, Jedrzejczak, R, Moy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2011-07-05 | | 公開日 | 2011-07-20 | | 最終更新日 | 2017-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.31 Å) | | 主引用文献 | Putative Mga family transcriptional regulator from Enterococcus faecalis.
To be Published
|
|
3S9X
 
 | | High resolution crystal structure of ASCH domain from Lactobacillus crispatus JV V101 | | 分子名称: | ASCH domain, CHLORIDE ION | | 著者 | Nocek, B, Xu, X, Cui, H, Jedrzejczak, R, Edwards, A, Savchenko, A, Mabbutt, B.C, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2011-06-02 | | 公開日 | 2011-07-27 | | 実験手法 | X-RAY DIFFRACTION (1.35 Å) | | 主引用文献 | High resolution crystal structure of ASCH domain from Lactobacillus crispatus JV V101
TO BE PUBLISHED
|
|
3SJR
 
 | |
3SBL
 
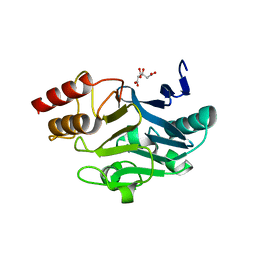 | | Crystal Structure of New Delhi Metal-beta-lactamase-1 from Klebsiella pneumoniae | | 分子名称: | Beta-lactamase NDM-1, CITRIC ACID | | 著者 | Kim, Y, Tesar, C, Jedrzejczak, R, Babnigg, J, Binkowski, T.A, Mire, J, Sacchettini, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Structures of Mtb Proteins Conferring Susceptibility to Known Mtb Inhibitors (MTBI) | | 登録日 | 2011-06-05 | | 公開日 | 2011-06-29 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.31 Å) | | 主引用文献 | Structure of Apo- and Monometalated Forms of NDM-1 A Highly Potent Carbapenem-Hydrolyzing Metallo-beta-Lactamase
Plos One, 6, 2011
|
|
3S4L
 
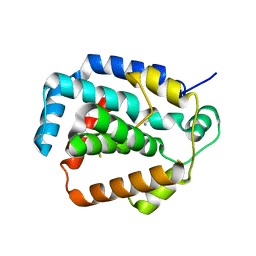 | | The CRISPR-associated Cas3 HD domain protein MJ0384 from Methanocaldococcus jannaschii | | 分子名称: | CALCIUM ION, CAS3 Metal dependent phosphohydrolase | | 著者 | Petit, P, Brown, G, Yakunin, A, Edwards, A, Joachimiak, A, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2011-05-19 | | 公開日 | 2011-06-22 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structure and activity of the Cas3 HD nuclease MJ0384, an effector enzyme of the CRISPR interference.
Embo J., 30, 2011
|
|
3TEB
 
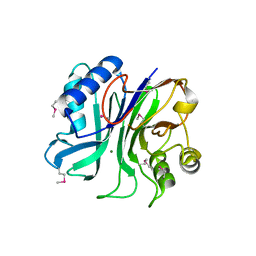 | | endonuclease/exonuclease/phosphatase family protein from Leptotrichia buccalis C-1013-b | | 分子名称: | Endonuclease/exonuclease/phosphatase, MAGNESIUM ION | | 著者 | Chang, C, Bigelow, L, Muniez, I, Bearden, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2011-08-12 | | 公開日 | 2011-08-31 | | 実験手法 | X-RAY DIFFRACTION (2.99 Å) | | 主引用文献 | Crystal structure of endonuclease/exonuclease/phosphatase family protein from Leptotrichia buccalis C-1013-b
To be Published
|
|
3T8K
 
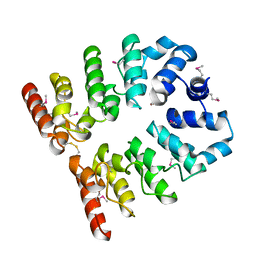 | |
3T9Y
 
 | | Crystal structure of GNAT family acetyltransferase Staphylococcus aureus subsp. aureus USA300_TCH1516 | | 分子名称: | 1,2-ETHANEDIOL, Acetyltransferase, GNAT family, ... | | 著者 | Chang, C, Tesar, C, Jedrzejczak, R, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2011-08-03 | | 公開日 | 2011-08-17 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystal structure of GNAT family acetyltransferase Staphylococcus aureus subsp. aureus USA300_TCH1516
To be Published
|
|
3TEV
 
 | | The crystal structure of glycosyl hydrolase from Deinococcus radiodurans R1 | | 分子名称: | Glycosyl hyrolase, family 3 | | 著者 | Chang, C, Hatzos-Skintges, C, Kohler, M, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2011-08-15 | | 公開日 | 2011-08-31 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | The crystal structure of glycosyl hydrolase from Deinococcus radiodurans R1
To be Published
|
|
3UO2
 
 | | Jac1 co-chaperone from Saccharomyces cerevisiae | | 分子名称: | J-type co-chaperone JAC1, mitochondrial | | 著者 | Osipiuk, J, Mulligan, R, Bigelow, L, Marszalek, J, Craig, E.A, Dutkiewicz, R, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2011-11-16 | | 公開日 | 2011-12-14 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.13 Å) | | 主引用文献 | Interaction of j-protein co-chaperone jac1 with fe-s scaffold isu is indispensable in vivo and conserved in evolution.
J.Mol.Biol., 417, 2012
|
|
3V75
 
 | | Crystal structure of putative orotidine 5'-phosphate decarboxylase from Streptomyces avermitilis ma-4680 | | 分子名称: | Orotidine 5'-phosphate decarboxylase | | 著者 | Stogios, P.J, Xu, X, Cui, H, Kudritska, M, Tan, K, Edwards, A, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2011-12-20 | | 公開日 | 2012-05-09 | | 最終更新日 | 2017-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Crystal structure of putative orotidine 5'-phosphate decarboxylase from Streptomyces avermitilis ma-4680
To be Published
|
|
3SG0
 
 | | The crystal structure of an extracellular ligand-binding receptor from Rhodopseudomonas palustris HaA2 | | 分子名称: | BENZOYL-FORMIC ACID, Extracellular ligand-binding receptor | | 著者 | Tan, K, Mack, J.C, Zerbs, S, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2011-06-14 | | 公開日 | 2011-06-29 | | 最終更新日 | 2015-10-14 | | 実験手法 | X-RAY DIFFRACTION (1.201 Å) | | 主引用文献 | Structural and functional characterization of solute binding proteins for aromatic compounds derived from lignin: p-coumaric acid and related aromatic acids.
Proteins, 81, 2013
|
|
3TVA
 
 | | Crystal Structure of Xylose isomerase domain protein from Planctomyces limnophilus | | 分子名称: | CHLORIDE ION, GLYCEROL, MAGNESIUM ION, ... | | 著者 | Kim, Y, Wu, R, Bearden, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2011-09-19 | | 公開日 | 2011-10-05 | | 実験手法 | X-RAY DIFFRACTION (2.148 Å) | | 主引用文献 | Crystal Structure of Xylose isomerase domain protein from Planctomyces limnophilus
To be Published
|
|
3UK0
 
 | | RPD_1889 protein, an extracellular ligand-binding receptor from Rhodopseudomonas palustris. | | 分子名称: | 1,2-ETHANEDIOL, 3-(4-HYDROXY-PHENYL)PYRUVIC ACID, Extracellular ligand-binding receptor, ... | | 著者 | Osipiuk, J, Mack, J, Zerbs, S, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2011-11-08 | | 公開日 | 2011-11-23 | | 最終更新日 | 2017-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.49 Å) | | 主引用文献 | Structural and functional characterization of solute binding proteins for aromatic compounds derived from lignin: p-Coumaric acid and related aromatic acids.
Proteins, 81, 2013
|
|
3V7B
 
 | | Dip2269 protein from corynebacterium diphtheriae | | 分子名称: | 1,2-ETHANEDIOL, Uncharacterized protein | | 著者 | Osipiuk, J, Duggan, E, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2011-12-20 | | 公開日 | 2012-01-11 | | 最終更新日 | 2017-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.743 Å) | | 主引用文献 | Dip2269 protein from corynebacterium diphtheriae.
To be Published
|
|
3U4Y
 
 | | The crystal structure of a functionally unknown protein (Dtox_1751) from Desulfotomaculum acetoxidans DSM 771. | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, uncharacterized protein | | 著者 | Tan, K, Bigelow, L, Bearden, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2011-10-10 | | 公開日 | 2011-10-26 | | 実験手法 | X-RAY DIFFRACTION (2.994 Å) | | 主引用文献 | The crystal structure of a functionally unknown protein (Dtox_1751) from Desulfotomaculum acetoxidans DSM 771.
To be Published
|
|
3U7V
 
 | |
3SOZ
 
 | | Cytoplasmic Protein STM1381 from Salmonella typhimurium LT2 | | 分子名称: | Cytoplasmic Protein STM1381, GLYCEROL | | 著者 | Joachimiak, A, Duke, N.E.C, Jedrzejczak, R, Li, H, Adkins, J, Brown, R, Midwest Center for Structural Genomics (MCSG), Program for the Characterization of Secreted Effector Proteins (PCSEP) | | 登録日 | 2011-06-30 | | 公開日 | 2011-08-03 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Cytoplasmic Protein STM1381 from Salmonella typhimurium LT2
To be Published
|
|
3UO3
 
 | | Jac1 co-chaperone from Saccharomyces cerevisiae, 5-182 clone | | 分子名称: | ACETATE ION, J-type co-chaperone JAC1, mitochondrial | | 著者 | Osipiuk, J, Bigelow, L, Mulligan, R, Feldmann, B, Babnigg, G, Marszalek, J, Craig, E.A, Dutkiewicz, R, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2011-11-16 | | 公開日 | 2011-12-14 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Interaction of j-protein co-chaperone jac1 with fe-s scaffold isu is indispensable in vivo and conserved in evolution.
J.Mol.Biol., 417, 2012
|
|
3V77
 
 | | Crystal structure of a putative fumarylacetoacetate isomerase/hydrolase from Oleispira antarctica | | 分子名称: | ACETATE ION, D(-)-TARTARIC ACID, Putative fumarylacetoacetate isomerase/hydrolase, ... | | 著者 | Stogios, P.J, Kagan, O, Di Leo, R, Bochkarev, A, Edwards, A.M, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2011-12-20 | | 公開日 | 2012-01-18 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Genome sequence and functional genomic analysis of the oil-degrading bacterium Oleispira antarctica.
Nat Commun, 4, 2013
|
|
3UKJ
 
 | | Crystal structure of extracellular ligand-binding receptor from Rhodopseudomonas palustris HaA2 | | 分子名称: | 3-(4-HYDROXY-PHENYL)PYRUVIC ACID, Extracellular ligand-binding receptor, GLYCEROL, ... | | 著者 | Chang, C, Mack, J, Zerbs, S, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2011-11-09 | | 公開日 | 2011-11-23 | | 最終更新日 | 2013-09-25 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Structural and functional characterization of solute binding proteins for aromatic compounds derived from lignin: p-Coumaric acid and related aromatic acids.
Proteins, 81, 2013
|
|
3VCR
 
 | | Crystal structure of a putative Kdpg (2-keto-3-deoxy-6-phosphogluconate) aldolase from Oleispira antarctica | | 分子名称: | PYRUVIC ACID, putative Kdpg (2-keto-3-deoxy-6-phosphogluconate) aldolase | | 著者 | Stogios, P.J, Kagan, O, Di Leo, R, Yim, V, Joachimiak, A, Edwards, A.M, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2012-01-04 | | 公開日 | 2012-01-18 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.84 Å) | | 主引用文献 | Genome sequence and functional genomic analysis of the oil-degrading bacterium Oleispira antarctica.
Nat Commun, 4, 2013
|
|
3TTG
 
 | | Crystal structure of putative aminomethyltransferase from Leptospirillum rubarum | | 分子名称: | CHLORIDE ION, Putative aminomethyltransferase | | 著者 | Michalska, K, Xu, X, Cui, H, Chin, S, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2011-09-14 | | 公開日 | 2011-10-12 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystal structure of putative aminomethyltransferase from Leptospirillum rubarum
TO BE PUBLISHED
|
|
3TYK
 
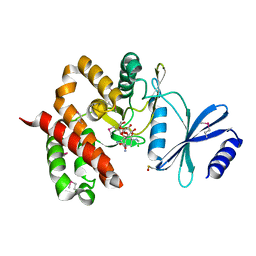 | | Crystal structure of aminoglycoside phosphotransferase APH(4)-Ia | | 分子名称: | CHLORIDE ION, HYGROMYCIN B VARIANT, Hygromycin-B 4-O-kinase | | 著者 | Stogios, P.J, Shabalin, I.G, Shakya, T, Evdokmova, E, Fan, Y, Chruszcz, M, Minor, W, Wright, G.D, Savchenko, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2011-09-26 | | 公開日 | 2011-10-12 | | 最終更新日 | 2022-04-13 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Structure and function of APH(4)-Ia, a hygromycin B resistance enzyme.
J.Biol.Chem., 286, 2011
|
|
