4GES
 
 | | crystal structure of GFP-TYR151PYZ with an unnatural amino acid incorporation | | 分子名称: | Green fluorescent protein | | 著者 | Dong, J, Liu, X, Li, J, Wang, J, Gong, W. | | 登録日 | 2012-08-02 | | 公開日 | 2012-08-29 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.23 Å) | | 主引用文献 | Genetic incorporation of a metal-chelating amino Acid as a probe for protein electron transfer.
Angew.Chem.Int.Ed.Engl., 51, 2012
|
|
3UN0
 
 | | Crystal Structure of MDC1 FHA Domain | | 分子名称: | Mediator of DNA damage checkpoint protein 1, SULFATE ION | | 著者 | Clapperton, J.A, Lloyd, J, Haire, L.F, Li, J, Smerdon, S.J. | | 登録日 | 2011-11-15 | | 公開日 | 2011-12-28 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | The molecular basis of ATM-dependent dimerization of the Mdc1 DNA damage checkpoint mediator.
Nucleic Acids Res., 40, 2012
|
|
1CRW
 
 | |
8ZCC
 
 | |
1ES9
 
 | | X-RAY CRYSTAL STRUCTURE OF R22K MUTANT OF THE MAMMALIAN BRAIN PLATELET-ACTIVATING FACTOR ACETYLHYDROLASES (PAF-AH) | | 分子名称: | PLATELET-ACTIVATING FACTOR ACETYLHYDROLASE IB GAMMA SUBUNIT | | 著者 | McMullen, T.W.P, Li, J, Sheffield, P.J, Aoki, J, Martin, T.W, Arai, H, Inoue, K, Derewenda, Z.S. | | 登録日 | 2000-04-07 | | 公開日 | 2000-05-31 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | The functional implications of the dimerization of the catalytic subunits of the mammalian brain platelet-activating factor acetylhydrolase (Ib).
Protein Eng., 13, 2000
|
|
3N2Y
 
 | |
7MUB
 
 | | KcsA Open gate E71V mutant in Potassium | | 分子名称: | Fab heavy chain, Fab light chain, POTASSIUM ION, ... | | 著者 | Rohaim, A, Li, J, Weingarth, M, Roux, B. | | 登録日 | 2021-05-14 | | 公開日 | 2022-03-23 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | A distinct mechanism of C-type inactivation in the Kv-like KcsA mutant E71V.
Nat Commun, 13, 2022
|
|
6KF5
 
 | | Microbial Hormone-sensitive lipase E53 mutant I256L | | 分子名称: | (4-nitrophenyl) hexanoate, 1,2-ETHANEDIOL, GLYCEROL, ... | | 著者 | Yang, X, Li, Z.Y, Li, J, Xu, X.W. | | 登録日 | 2019-07-06 | | 公開日 | 2020-07-08 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.09 Å) | | 主引用文献 | Microbial Hormone-sensitive lipase E53 mutant I256L
To Be Published
|
|
2LXH
 
 | | NMR structure of the RING domain in ubiquitin ligase gp78 | | 分子名称: | E3 ubiquitin-protein ligase AMFR, ZINC ION | | 著者 | Das, R, Linag, Y, Mariano, J, Li, J, Huang, T, King, A, Weissman, A, Ji, X, Byrd, R. | | 登録日 | 2012-08-27 | | 公開日 | 2013-08-28 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Allosteric regulation of E2:E3 interactions promote a processive ubiquitination machine.
Embo J., 32, 2013
|
|
3H84
 
 | | Crystal structure of GET3 | | 分子名称: | ATPase GET3, CHLORIDE ION, MAGNESIUM ION, ... | | 著者 | Hu, J, Li, J, Qian, X, Sha, B. | | 登録日 | 2009-04-28 | | 公開日 | 2009-12-22 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | The crystal structures of yeast Get3 suggest a mechanism for tail-anchored protein membrane insertion.
Plos One, 4, 2009
|
|
1MI1
 
 | | Crystal Structure of the PH-BEACH Domain of Human Neurobeachin | | 分子名称: | Neurobeachin | | 著者 | Jogl, G, Shen, Y, Gebauer, D, Li, J, Wiegmann, K, Kashkar, H, Kroenke, M, Tong, L, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2002-08-21 | | 公開日 | 2002-09-27 | | 最終更新日 | 2024-11-20 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Crystal structure of the BEACH domain reveals an unusual fold and extensive association with a novel PH domain.
EMBO J., 21, 2002
|
|
4LRH
 
 | | Crystal structure of human folate receptor alpha in complex with folic acid | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, FOLIC ACID, Folate receptor alpha | | 著者 | Ke, J, Chen, C, Zhou, X.E, Yi, W, Brunzelle, J.S, Li, J, Young, E.-L, Xu, H.E, Melcher, K. | | 登録日 | 2013-07-19 | | 公開日 | 2013-07-31 | | 最終更新日 | 2024-11-27 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Structural basis for molecular recognition of folic acid by folate receptors.
Nature, 500, 2013
|
|
1CI5
 
 | | GLYCAN-FREE MUTANT ADHESION DOMAIN OF HUMAN CD58 (LFA-3) | | 分子名称: | PROTEIN (LYMPHOCYTE FUNCTION-ASSOCIATED ANTIGEN 3(CD58)) | | 著者 | Sun, Z.Y.J, Dotsch, V, Kim, M, Li, J, Reinherz, E.L, Wagner, G. | | 登録日 | 1999-04-07 | | 公開日 | 1999-06-22 | | 最終更新日 | 2023-12-27 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Functional glycan-free adhesion domain of human cell surface receptor CD58: design, production and NMR studies.
EMBO J., 18, 1999
|
|
6P49
 
 | | Cryo-EM structure of calcium-bound TMEM16F in nanodisc with supplement of PIP2 in Cl2 | | 分子名称: | Anoctamin-6, CALCIUM ION | | 著者 | Feng, S, Dang, S, Han, T.W, Ye, W, Jin, P, Cheng, T, Li, J, Jan, Y.N, Jan, L.Y, Cheng, Y. | | 登録日 | 2019-05-26 | | 公開日 | 2019-07-24 | | 最終更新日 | 2024-10-09 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Cryo-EM Studies of TMEM16F Calcium-Activated Ion Channel Suggest Features Important for Lipid Scrambling.
Cell Rep, 28, 2019
|
|
6P46
 
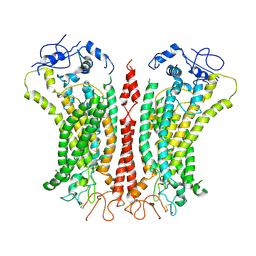 | | Cryo-EM structure of TMEM16F in digitonin with calcium bound | | 分子名称: | Anoctamin-6, CALCIUM ION | | 著者 | Feng, S, Dang, S, Han, T.W, Ye, W, Jin, P, Cheng, T, Li, J, Jan, Y.N, Jan, L.Y, Cheng, Y. | | 登録日 | 2019-05-26 | | 公開日 | 2019-07-24 | | 最終更新日 | 2024-11-20 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | Cryo-EM Studies of TMEM16F Calcium-Activated Ion Channel Suggest Features Important for Lipid Scrambling.
Cell Rep, 28, 2019
|
|
6P47
 
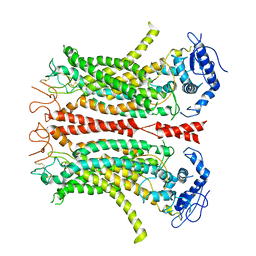 | | Cryo-EM structure of TMEM16F in digitonin without calcium | | 分子名称: | Anoctamin-6 | | 著者 | Feng, S, Dang, S, Han, T.W, Ye, W, Jin, P, Cheng, T, Li, J, Jan, Y.N, Jan, L.Y, Cheng, Y. | | 登録日 | 2019-05-26 | | 公開日 | 2019-07-24 | | 最終更新日 | 2024-11-20 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | Cryo-EM Studies of TMEM16F Calcium-Activated Ion Channel Suggest Features Important for Lipid Scrambling.
Cell Rep, 28, 2019
|
|
8WUR
 
 | | Crystal structure of SARS-Cov-2 main protease D48N mutant in complex with shikonin | | 分子名称: | 2-[(1R)-4-methyl-1-oxidanyl-pent-3-enyl]-5,8-bis(oxidanyl)naphthalene-1,4-dione, 3C-like proteinase nsp5 | | 著者 | Zhao, Z.Y, Li, W.W, Zhang, J, Li, J. | | 登録日 | 2023-10-21 | | 公開日 | 2024-11-13 | | 実験手法 | X-RAY DIFFRACTION (2.08 Å) | | 主引用文献 | Structural Basis for the Inhibition of SARS-CoV-2 M pro D48N Mutant by Shikonin and PF-07321332.
Viruses, 16, 2023
|
|
6P48
 
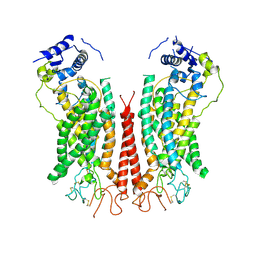 | | Cryo-EM structure of calcium-bound TMEM16F in nanodisc with supplement of PIP2 in Cl1 | | 分子名称: | Anoctamin-6, CALCIUM ION | | 著者 | Feng, S, Dang, S, Han, T.W, Ye, W, Jin, P, Cheng, T, Li, J, Jan, Y.N, Jan, L.Y, Cheng, Y. | | 登録日 | 2019-05-26 | | 公開日 | 2019-07-24 | | 最終更新日 | 2024-11-06 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Cryo-EM Studies of TMEM16F Calcium-Activated Ion Channel Suggest Features Important for Lipid Scrambling.
Cell Rep, 28, 2019
|
|
1WUG
 
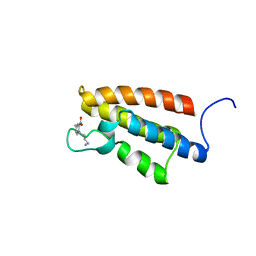 | | complex structure of PCAF bromodomain with small chemical ligand NP1 | | 分子名称: | Histone acetyltransferase PCAF, N-(3-AMINOPROPYL)-4-METHYL-2-NITROBENZENAMINE | | 著者 | Zeng, L, Li, J, Muller, M, Yan, S, Mujtaba, S, Pan, C, Wang, Z, Zhou, M.M. | | 登録日 | 2004-12-07 | | 公開日 | 2005-08-16 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Selective small molecules blocking HIV-1 Tat and coactivator PCAF association
J.Am.Chem.Soc., 127, 2005
|
|
1WUM
 
 | | Complex structure of PCAF bromodomain with small chemical ligand NP2 | | 分子名称: | Histone acetyltransferase PCAF, N-(3-AMINOPROPYL)-2-NITROBENZENAMINE | | 著者 | Zeng, L, Li, J, Muller, M, Yan, S, Mujtaba, S, Pan, C, Wang, Z, Zhou, M.M. | | 登録日 | 2004-12-08 | | 公開日 | 2005-08-16 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Selective small molecules blocking HIV-1 Tat and coactivator PCAF association
J.Am.Chem.Soc., 127, 2005
|
|
4MYS
 
 | | 1.4 Angstrom Crystal Structure of 2-succinyl-6-hydroxy-2,4-cyclohexadiene-1-carboxylate synthase with SHCHC and Pyruvate | | 分子名称: | 2-(3-CARBOXYPROPIONYL)-6-HYDROXY-CYCLOHEXA-2,4-DIENE CARBOXYLIC ACID, 2-succinyl-6-hydroxy-2,4-cyclohexadiene-1-carboxylate synthase, GLYCEROL, ... | | 著者 | Sun, Y, Yin, S, Feng, Y, Li, J, Zhou, J, Liu, C, Zhu, G, Guo, Z. | | 登録日 | 2013-09-28 | | 公開日 | 2014-04-23 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.423 Å) | | 主引用文献 | Molecular basis of the general base catalysis of an alpha / beta-hydrolase catalytic triad.
J.Biol.Chem., 289, 2014
|
|
4MXD
 
 | | 1.45 angstronm crystal structure of E.coli 2-succinyl-6-hydroxy-2,4-cyclohexadiene-1-carboxylate synthase (MenH) | | 分子名称: | 2-succinyl-6-hydroxy-2,4-cyclohexadiene-1-carboxylate synthase, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | 著者 | Sun, Y, Yin, S, Feng, Y, Li, J, Zhou, J, Liu, C, Zhu, G, Guo, Z. | | 登録日 | 2013-09-26 | | 公開日 | 2014-04-23 | | 最終更新日 | 2024-10-09 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | Molecular basis of the general base catalysis of an alpha / beta-hydrolase catalytic triad.
J.Biol.Chem., 289, 2014
|
|
4PT1
 
 | |
4NXE
 
 | | Crystal structure of iLOV-I486(2LT) at pH 6.5 | | 分子名称: | FLAVIN MONONUCLEOTIDE, Phototropin-2 | | 著者 | Wang, J, Liu, X, Li, J. | | 登録日 | 2013-12-09 | | 公開日 | 2014-09-24 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.103 Å) | | 主引用文献 | Significant expansion of fluorescent protein sensing ability through the genetic incorporation of superior photo-induced electron-transfer quenchers.
J.Am.Chem.Soc., 136, 2014
|
|
8WF7
 
 | | The Crystal Structure of integrase from Biortus | | 分子名称: | ACETATE ION, Integrase, SULFATE ION | | 著者 | Wang, F, Cheng, W, Yuan, Z, Qi, J, Li, J. | | 登録日 | 2023-09-19 | | 公開日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | The Crystal Structure of integrase from Biortus
To Be Published
|
|
