8OYV
 
 | |
8OYS
 
 | |
7JZN
 
 | |
7JZM
 
 | |
7UE2
 
 | |
7UDK
 
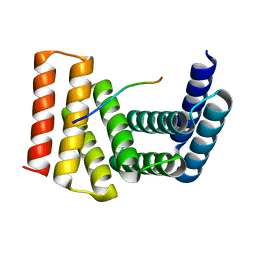 | | Crystal structure of designed helical repeat protein RPB_LRP2_R4 bound to LRPx4 peptide | | 分子名称: | 4xLRP, Designed helical repeat protein (DHR) RPB_LRP2_R4 | | 著者 | Chang, Y, Redler, R.L, Bhabha, G, Ekiert, D.C. | | 登録日 | 2022-03-20 | | 公開日 | 2023-03-22 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (3.18 Å) | | 主引用文献 | De novo design of modular peptide-binding proteins by superhelical matching.
Nature, 616, 2023
|
|
7UDO
 
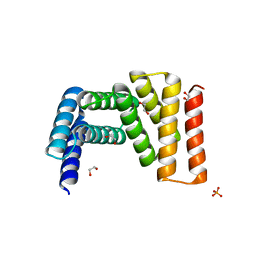 | | Crystal structure of designed helical repeat protein RPB_LRP2_R4 (proteolysis fragment?), forming pseudopolymeric filaments | | 分子名称: | 1,2-ETHANEDIOL, Designed helical repeat protein (DHR) RPB_LRP2_R4, PHOSPHATE ION | | 著者 | Redler, R.L, Chang, Y, Bhabha, G, Ekiert, D.C. | | 登録日 | 2022-03-20 | | 公開日 | 2023-03-22 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | De novo design of modular peptide-binding proteins by superhelical matching.
Nature, 616, 2023
|
|
7UDL
 
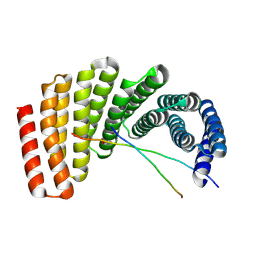 | | Crystal structure of designed helical repeat protein RPB_PLP1_R6 bound to PLPx6 peptide | | 分子名称: | 1,2-ETHANEDIOL, 6xPLP Peptide, Designed helical repeat protein (DHR) RPB_PLP1_R6 | | 著者 | Chang, Y, Redler, R.L, Bhabha, G, Ekiert, D.C. | | 登録日 | 2022-03-20 | | 公開日 | 2023-03-22 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | De novo design of modular peptide-binding proteins by superhelical matching.
Nature, 616, 2023
|
|
7UDN
 
 | |
7UDM
 
 | | Crystal structure of designed helical repeat protein RPB_PLP1_R6 in alternative conformation 1 (with peptide) | | 分子名称: | 6xPLP, Designed helical repeat protein (DHR) RPB_PLP1_R6 | | 著者 | Chang, Y, Redler, R.L, Bhabha, G, Ekiert, D.C. | | 登録日 | 2022-03-20 | | 公開日 | 2023-03-22 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.65 Å) | | 主引用文献 | De novo design of modular peptide-binding proteins by superhelical matching.
Nature, 616, 2023
|
|
7JZU
 
 | |
3I1C
 
 | |
3IIV
 
 | |
2N44
 
 | | EC-NMR Structure of Escherichia coli Maltose-binding protein Determined by Combining Evolutionary Couplings (EC) and Sparse NMR Data. Northeast Structural Genomics Consortium target ER690 | | 分子名称: | Maltose-binding periplasmic protein | | 著者 | Tang, Y, Huang, Y.J, Hopf, T.A, Sander, C, Marks, D, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2015-06-16 | | 公開日 | 2015-07-01 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Protein structure determination by combining sparse NMR data with evolutionary couplings.
Nat.Methods, 12, 2015
|
|
2N45
 
 | | EC-NMR Structure of Escherichia coli Maltose-binding protein Determined by Combining Evolutionary Couplings (EC) and Sparse NMR Data with a second set of RDC data simulated for an alternative alignment tensor. Northeast Structural Genomics Consortium target ER690 | | 分子名称: | Maltose-binding periplasmic protein | | 著者 | Tang, Y, Huang, Y.J, Hopf, T.A, Sander, C, Marks, D, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2015-06-17 | | 公開日 | 2015-07-01 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Protein structure determination by combining sparse NMR data with evolutionary couplings.
Nat.Methods, 12, 2015
|
|
2N47
 
 | | EC-NMR Structure of Synechocystis sp. PCC 6803 Slr1183 Determined by Combining Evolutionary Couplings (EC) and Sparse NMR Data. Northeast Structural Genomics Consortium target SgR145 | | 分子名称: | Slr1183 protein | | 著者 | Tang, Y, Huang, Y.J, Hopf, T.A, Sander, C, Marks, D, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2015-06-17 | | 公開日 | 2015-07-01 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Protein structure determination by combining sparse NMR data with evolutionary couplings.
Nat.Methods, 12, 2015
|
|
5VL4
 
 | | Accidental minimum contact crystal lattice formed by a redesigned protein oligomer | | 分子名称: | T33-53H-B | | 著者 | Cannon, K.A, Cascio, D, Sawaya, M.R, Park, R, Boyken, S, King, N, Yeates, T. | | 登録日 | 2017-04-24 | | 公開日 | 2018-05-23 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (4.1 Å) | | 主引用文献 | Design and structure of two new protein cages illustrate successes and ongoing challenges in protein engineering.
Protein Sci., 29, 2020
|
|
3IIO
 
 | |
3IIP
 
 | |
6DG4
 
 | |
2LAF
 
 | |
6MOH
 
 | | Dimeric DARPin C_R3 complex with EpoR | | 分子名称: | Dimeric DARPin E2C (C_R3), Erythropoietin receptor, PHOSPHATE ION, ... | | 著者 | Jude, K.M, Mohan, K, Garcia, K.C. | | 登録日 | 2018-10-04 | | 公開日 | 2019-06-05 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | Topological control of cytokine receptor signaling induces differential effects in hematopoiesis.
Science, 364, 2019
|
|
6MOI
 
 | | Dimeric DARPin C_angle_R5 complex with EpoR | | 分子名称: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Dimeric DARPing CCR5 (C_angle_R5), ... | | 著者 | Jude, K.M, Mohan, K, Garcia, K.C, Guo, Y. | | 登録日 | 2018-10-04 | | 公開日 | 2019-06-05 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.065 Å) | | 主引用文献 | Topological control of cytokine receptor signaling induces differential effects in hematopoiesis.
Science, 364, 2019
|
|
6MOG
 
 | | Dimeric DARPin C_R3 | | 分子名称: | 1,2-ETHANEDIOL, DARPin C_R3, TRIETHYLENE GLYCOL | | 著者 | Jude, K.M, Mohan, K, Garcia, K.C. | | 登録日 | 2018-10-04 | | 公開日 | 2019-06-05 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.21 Å) | | 主引用文献 | Topological control of cytokine receptor signaling induces differential effects in hematopoiesis.
Science, 364, 2019
|
|
6MOK
 
 | | Dimeric DARPin A_distance_R7 complex with EpoR | | 分子名称: | Dimeric DARPin ANR7 (A_distance_r7), Erythropoietin receptor | | 著者 | Jude, K.M, Mohan, K, Garcia, K.C, Guo, Y. | | 登録日 | 2018-10-04 | | 公開日 | 2019-06-05 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (5.101 Å) | | 主引用文献 | Topological control of cytokine receptor signaling induces differential effects in hematopoiesis.
Science, 364, 2019
|
|
