4FFV
 
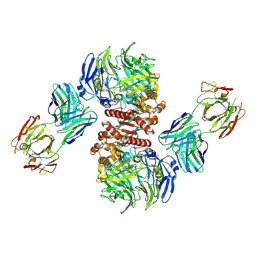 | | Crystal Structure of Dipeptidyl Peptidase IV (DPP4, DPP-IV, CD26) in Complex with 11A19 Fab | | 分子名称: | 11A19 Fab heavy chain, 11A19 Fab light chain, Dipeptidyl peptidase 4 | | 著者 | Wang, Z, Sudom, A, Walker, N.P, Min, X. | | 登録日 | 2012-06-01 | | 公開日 | 2012-12-12 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | An Inhibitory Antibody Against DPP IV Improves Glucose Tolerance in vivo - Validation of Large Molecule Approach for DPP IV Inhibition
To be published
|
|
4FFW
 
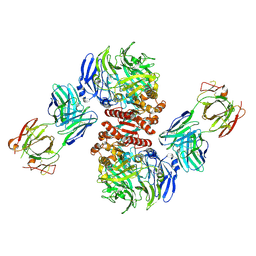 | | Crystal Structure of Dipeptidyl Peptidase IV (DPP4, DPP-IV, CD26) in Complex with Fab + sitagliptin | | 分子名称: | (2R)-4-OXO-4-[3-(TRIFLUOROMETHYL)-5,6-DIHYDRO[1,2,4]TRIAZOLO[4,3-A]PYRAZIN-7(8H)-YL]-1-(2,4,5-TRIFLUOROPHENYL)BUTAN-2-A MINE, Dipeptidyl peptidase 4, Fab heavy chain, ... | | 著者 | Wang, Z, Sudom, A, Walker, N.P, Min, X. | | 登録日 | 2012-06-01 | | 公開日 | 2012-12-12 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | An Inhibitory Antibody Against DPP IV Improves Glucose Tolerance in vivo - Validation of Large Molecule Approach for DPP IV Inhibition
To be published
|
|
8CO3
 
 | |
8CO0
 
 | |
3QGT
 
 | | Crystal structure of Wild-type PfDHFR-TS COMPLEXED WITH NADPH, dUMP AND PYRIMETHAMINE | | 分子名称: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, 5-(4-CHLORO-PHENYL)-6-ETHYL-PYRIMIDINE-2,4-DIAMINE, Bifunctional dihydrofolate reductase-thymidylate synthase, ... | | 著者 | Chitnumsub, P, Yuthavong, Y. | | 登録日 | 2011-01-24 | | 公開日 | 2011-06-29 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Trypanosomal dihydrofolate reductase reveals natural antifolate resistance
Acs Chem.Biol., 6, 2011
|
|
9JH6
 
 | | Activation mechanism of CYSLTR2 by C20:0 | | 分子名称: | Cer(d18:0/20:0), Cysteinyl leukotriene receptor 2, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | 著者 | Wang, J.L, Sun, J.P, Yu, X. | | 登録日 | 2024-09-09 | | 公開日 | 2025-04-30 | | 最終更新日 | 2025-05-21 | | 実験手法 | ELECTRON MICROSCOPY (2.89 Å) | | 主引用文献 | Sensing ceramides by CYSLTR2 and P2RY6 to aggravate atherosclerosis.
Nature, 641, 2025
|
|
6EOY
 
 | |
8D36
 
 | |
8D6Z
 
 | | Crystal structure of SARS-CoV-2 spike stem fusion peptide in complex with neutralizing antibody COV91-27 | | 分子名称: | Neutralizing antibody COV91-27 heavy chain, Neutralizing antibody COV91-27 light chain, Spike protein S2 fusion peptide | | 著者 | Lee, C.C.D, Lin, T.H, Yuan, M, Wilson, I.A. | | 登録日 | 2022-06-06 | | 公開日 | 2022-07-27 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Broadly neutralizing antibodies target the coronavirus fusion peptide.
Science, 377, 2022
|
|
6T81
 
 | | Human Carbonic anhydrase II bound by 2-Naphthalenesulfonamide. | | 分子名称: | AZIDE ION, BICINE, SODIUM ION, ... | | 著者 | Smirnov, A, Manakova, E, Grazulis, S. | | 登録日 | 2019-10-23 | | 公開日 | 2020-10-14 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (0.98 Å) | | 主引用文献 | Isoform-Selective Enzyme Inhibitors by Exploring Pocket Size According to the Lock-and-Key Principle.
Biophys.J., 119, 2020
|
|
6T4N
 
 | | Human Carbonic anhydrase II bound by 2,4,6-trimethylbenzenesulfonamide | | 分子名称: | 2,4,6-trimethylbenzenesulfonamide, 2-piperazin-1-ylethanol, BICINE, ... | | 著者 | Smirnov, A, Manakova, E, Grazulis, S. | | 登録日 | 2019-10-14 | | 公開日 | 2020-10-14 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Isoform-Selective Enzyme Inhibitors by Exploring Pocket Size According to the Lock-and-Key Principle.
Biophys.J., 119, 2020
|
|
6T5C
 
 | | Human Carbonic anhydrase II bound by anthracene-9-sulfonamide | | 分子名称: | BICINE, DI(HYDROXYETHYL)ETHER, DIMETHYL SULFOXIDE, ... | | 著者 | Smirnov, A, Manakova, E, Grazulis, S. | | 登録日 | 2019-10-15 | | 公開日 | 2020-10-14 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.22 Å) | | 主引用文献 | Isoform-Selective Enzyme Inhibitors by Exploring Pocket Size According to the Lock-and-Key Principle.
Biophys.J., 119, 2020
|
|
6T5Q
 
 | | Human Carbonic anhydrase XII bound by 3,5-diphenylbenzenesulfonamide | | 分子名称: | 1,2-ETHANEDIOL, 3,5-diphenylbenzenesulfonamide, Carbonic anhydrase 12, ... | | 著者 | Smirnov, A, Manakova, E, Grazulis, S. | | 登録日 | 2019-10-16 | | 公開日 | 2020-10-14 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Isoform-Selective Enzyme Inhibitors by Exploring Pocket Size According to the Lock-and-Key Principle.
Biophys.J., 119, 2020
|
|
3E4H
 
 | | Crystal structure of the cyclotide varv F | | 分子名称: | Varv peptide F,Varv peptide F | | 著者 | Hu, S.H. | | 登録日 | 2008-08-11 | | 公開日 | 2009-02-10 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Combined X-ray and NMR Analysis of the Stability of the Cyclotide Cystine Knot Fold That Underpins Its Insecticidal Activity and Potential Use as a Drug Scaffold
J.Biol.Chem., 284, 2009
|
|
3LQ8
 
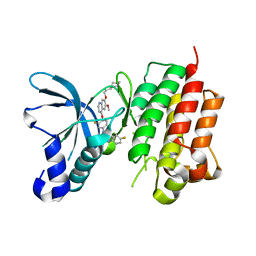 | | Structure of the kinase domain of c-Met bound to XL880 (GSK1363089) | | 分子名称: | Hepatocyte growth factor receptor, N-(3-fluoro-4-{[6-methoxy-7-(3-morpholin-4-ylpropoxy)quinolin-4-yl]oxy}phenyl)-N'-(4-fluorophenyl)cyclopropane-1,1-dicarboxamide | | 著者 | Lougheed, J.C, Stout, T.J. | | 登録日 | 2010-02-08 | | 公開日 | 2010-05-19 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.02 Å) | | 主引用文献 | Inhibition of tumor cell growth, invasion, and metastasis by EXEL-2880 (XL880, GSK1363089), a novel inhibitor of HGF and VEGF receptor tyrosine kinases.
Cancer Res., 69, 2009
|
|
8XEP
 
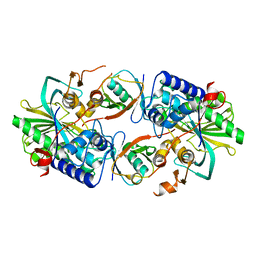 | | Crystal structure of a Legionella pneumophila type IV effector in complex with ubiquitin | | 分子名称: | SULFATE ION, Type IV effector MavL, Ubiquitin | | 著者 | Tan, J.X, Wang, X.F, Zhou, Y, Zhu, Y.Q. | | 登録日 | 2023-12-12 | | 公開日 | 2024-05-01 | | 最終更新日 | 2024-07-24 | | 実験手法 | X-RAY DIFFRACTION (2.95 Å) | | 主引用文献 | Legionella effector LnaB is a phosphoryl AMPylase that impairs phosphosignalling.
Nature, 631, 2024
|
|
8PWW
 
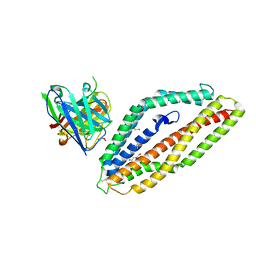 | |
8PWU
 
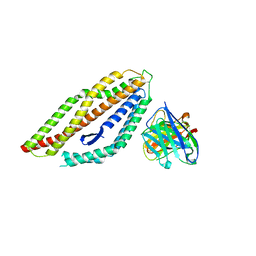 | | PfRH5 bound to monoclonal antibody MAD10-255 | | 分子名称: | Reticulocyte-binding protein homolog 5, monoclonal antibody MAD10-255 | | 著者 | Farrell, B, Higgins, M.K. | | 登録日 | 2023-07-21 | | 公開日 | 2024-06-05 | | 最終更新日 | 2024-12-18 | | 実験手法 | X-RAY DIFFRACTION (3.148 Å) | | 主引用文献 | Natural malaria infection elicits rare but potent neutralizing antibodies to the blood-stage antigen RH5.
Cell, 187, 2024
|
|
8Q5D
 
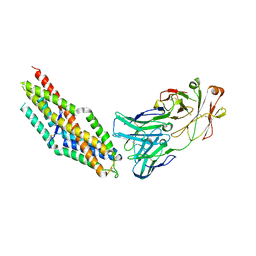 | | PfRH5 bound to monoclonal antibody MAD10-466 | | 分子名称: | Monoclonal antibody MAD10-466 heavy chain, Monoclonal antibody MAD10-466 light chain, Reticulocyte-binding protein homolog 5 | | 著者 | Farrell, B, Higgins, M.K. | | 登録日 | 2023-08-08 | | 公開日 | 2024-06-05 | | 最終更新日 | 2024-12-18 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | Natural malaria infection elicits rare but potent neutralizing antibodies to the blood-stage antigen RH5.
Cell, 187, 2024
|
|
8PWV
 
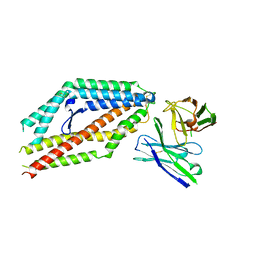 | | PfRH5 bound to monoclonal antibody MAD8-502 | | 分子名称: | Reticulocyte-binding protein homolog 5, monoclonal antibody MAD8-502 | | 著者 | Farrell, B, Higgins, M.K. | | 登録日 | 2023-07-21 | | 公開日 | 2024-06-05 | | 最終更新日 | 2024-12-18 | | 実験手法 | X-RAY DIFFRACTION (2.07 Å) | | 主引用文献 | Natural malaria infection elicits rare but potent neutralizing antibodies to the blood-stage antigen RH5.
Cell, 187, 2024
|
|
8PWX
 
 | | PfRH5 bound to monoclonal antibody R5.008 | | 分子名称: | GLYCEROL, Reticulocyte-binding protein homolog 5, scFv fragment for antibody R5.008 | | 著者 | Farrell, B, Higgins, M.K. | | 登録日 | 2023-07-21 | | 公開日 | 2024-06-05 | | 最終更新日 | 2024-12-18 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | Natural malaria infection elicits rare but potent neutralizing antibodies to the blood-stage antigen RH5.
Cell, 187, 2024
|
|
8QKS
 
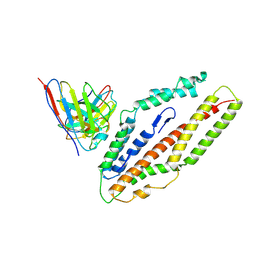 | | Plasmodium falciparum reticulocyte-binding protein homologue 5 (PfRH5) bound to R5.034 | | 分子名称: | Immunoglobulin lambda variable 1-36, R5034HV, Reticulocyte-binding protein-like protein 5 | | 著者 | Wright, N.D, Barrett, J.R, Bradshaw, W.J, Paterson, N.G, MacLean, E.M, Ferreira, L, McHugh, K, Von Delft, F, Koekemoer, L, Draper, S.J. | | 登録日 | 2023-09-16 | | 公開日 | 2024-07-31 | | 最終更新日 | 2024-09-18 | | 実験手法 | X-RAY DIFFRACTION (3.994 Å) | | 主引用文献 | Analysis of the diverse antigenic landscape of the malaria protein RH5 identifies a potent vaccine-induced human public antibody clonotype.
Cell, 187, 2024
|
|
8QKR
 
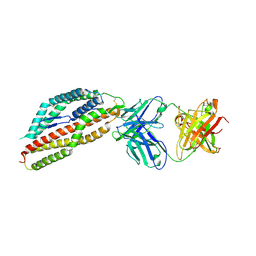 | | Plasmodium falciparum reticulocyte-binding protein homologue 5 (PfRH5) bound to R5.251 | | 分子名称: | R5251VHCH, R5251VLCL, Reticulocyte-binding protein-like protein 5 | | 著者 | Wright, N.D, Barrett, J.R, Bradshaw, W.J, Paterson, N.G, MacLean, E.M, Ferreira, L, McHugh, K, Koekemoer, L, Draper, S.J. | | 登録日 | 2023-09-16 | | 公開日 | 2024-07-31 | | 最終更新日 | 2024-11-13 | | 実験手法 | X-RAY DIFFRACTION (3.234 Å) | | 主引用文献 | Analysis of the diverse antigenic landscape of the malaria protein RH5 identifies a potent vaccine-induced human public antibody clonotype.
Cell, 187, 2024
|
|
9JQL
 
 | |
8S5I
 
 | |
