6IVZ
 
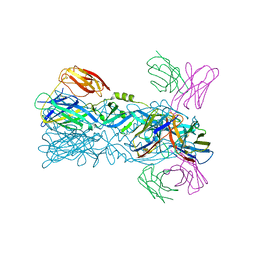 | | Crystal structure of 5A ScFv complexed with YFV-China sE in postfusion state | | 分子名称: | Envelope protein, Heavy chain of monoclonal antibody 5A, Light chain of monoclonal antibody 5A | | 著者 | Lu, X.S, Xiao, H.X, Li, S.H, Pang, X.F. | | 登録日 | 2018-12-04 | | 公開日 | 2019-02-13 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Double Lock of a Human Neutralizing and Protective Monoclonal Antibody Targeting the Yellow Fever Virus Envelope.
Cell Rep, 26, 2019
|
|
6IW4
 
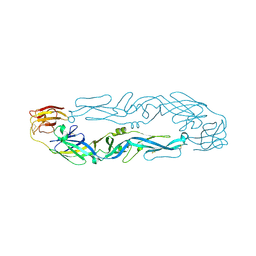 | |
6IW0
 
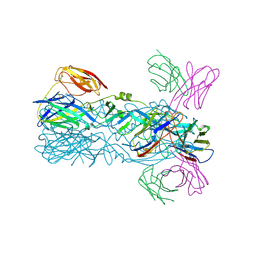 | | Crystal structure of 5A ScFv in complex with YFV-17D sE in postfusion state | | 分子名称: | Envelope protein E, Heavy chain of monoclonal antibody 5A, Light chain of monoclonal antibody 5A | | 著者 | Lu, X.S, Xiao, H.X, Li, S.H, Pang, X.F. | | 登録日 | 2018-12-04 | | 公開日 | 2019-02-13 | | 最終更新日 | 2019-02-27 | | 実験手法 | X-RAY DIFFRACTION (3.6 Å) | | 主引用文献 | Double Lock of a Human Neutralizing and Protective Monoclonal Antibody Targeting the Yellow Fever Virus Envelope.
Cell Rep, 26, 2019
|
|
6IW1
 
 | |
6IYF
 
 | | Structure of pSTING complex | | 分子名称: | (2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-2,9-bis(6-amino-9H-purin-9-yl)octahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8 ]tetraoxadiphosphacyclododecine-3,5,10,12-tetrol 5,12-dioxide, SULFATE ION, Stimulator of interferon genes protein | | 著者 | Yuan, Z.L, Shang, G.J, Cong, X.Y, Gu, L.C. | | 登録日 | 2018-12-15 | | 公開日 | 2019-06-19 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (1.764 Å) | | 主引用文献 | Crystal structures of porcine STINGCBD-CDN complexes reveal the mechanism of ligand recognition and discrimination of STING proteins.
J.Biol.Chem., 294, 2019
|
|
4ER7
 
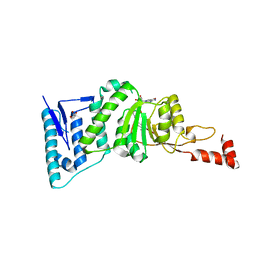 | | Crystal Structure of human DOT1L in complex with inhibitor SGC0947 | | 分子名称: | 5-bromo-7-{5-[(3-{[(4-tert-butylphenyl)carbamoyl]amino}propyl)amino]-5-deoxy-beta-D-ribofuranosyl}-7H-pyrrolo[2,3-d]pyrimidin-4-amine, GLYCEROL, Histone-lysine N-methyltransferase, ... | | 著者 | Wernimont, A.K, Tempel, W, Yu, W, Scopton, A, Li, Y, Nguyen, K.T, Vedadi, M, Bradner, J.E, Schapira, M, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Brown, P.J, Structural Genomics Consortium (SGC) | | 登録日 | 2012-04-19 | | 公開日 | 2012-05-16 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Catalytic site remodelling of the DOT1L methyltransferase by selective inhibitors.
Nat Commun, 3, 2012
|
|
4ER6
 
 | | Crystal structure of human DOT1L in complex with inhibitor SGC0946 | | 分子名称: | 5-bromo-7-{5-[(3-{[(4-tert-butylphenyl)carbamoyl]amino}propyl)(propan-2-yl)amino]-5-deoxy-beta-D-ribofuranosyl}-7H-pyrrolo[2,3-d]pyrimidin-4-amine, BROMIDE ION, Histone-lysine N-methyltransferase, ... | | 著者 | Wernimont, A.K, Tempel, W, Yu, W, Scopton, A, Li, Y, Nguyen, K.T, Vedadi, M, Bradner, J.E, Schapira, M, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Brown, P.J, Structural Genomics Consortium (SGC) | | 登録日 | 2012-04-19 | | 公開日 | 2012-05-16 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Catalytic site remodelling of the DOT1L methyltransferase by selective inhibitors.
Nat Commun, 3, 2012
|
|
8XG2
 
 | | The structure of HLA-A/Pep14 | | 分子名称: | Beta-2-microglobulin, HLA class I heavy chain, Spike protein S1 | | 著者 | Zhang, J.N, Yue, C, Liu, J. | | 登録日 | 2023-12-14 | | 公開日 | 2024-07-10 | | 実験手法 | X-RAY DIFFRACTION (1.84 Å) | | 主引用文献 | Uncommon P1 Anchor-featured Viral T Cell Epitope Preference within HLA-A*2601 and HLA-A*0101 Individuals.
Immunohorizons, 8, 2024
|
|
8X85
 
 | | Structure of leptin-LepR dimer | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Leptin, Leptin receptor, ... | | 著者 | Xie, Y.F, Shang, G.J, Qi, J.X, Gao, G.F. | | 登録日 | 2023-11-27 | | 公開日 | 2024-08-07 | | 実験手法 | ELECTRON MICROSCOPY (3.58 Å) | | 主引用文献 | Structural plasticity of human leptin binding to its receptor LepR
Hlife, 1, 2023
|
|
8XES
 
 | | The structure of HLA-A/L1-1 | | 分子名称: | Beta-2-microglobulin, HLA class I heavy chain, Major capsid protein L1 | | 著者 | Zhang, J.N, Yue, C, Liu, J. | | 登録日 | 2023-12-12 | | 公開日 | 2024-07-10 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (1.78 Å) | | 主引用文献 | Uncommon P1 Anchor-featured Viral T Cell Epitope Preference within HLA-A*2601 and HLA-A*0101 Individuals.
Immunohorizons, 8, 2024
|
|
8WU3
 
 | |
8XKE
 
 | | The structure of HLA-A/14-3-D | | 分子名称: | Beta-2-microglobulin, GLU-VAL-ASP-ASN-ALA-THR-ARG-PHE-ALA-SER-VAL-TYR, HLA class I heavy chain | | 著者 | Zhang, J.N, Yue, C, Liu, J, Sun, Z.Y. | | 登録日 | 2023-12-23 | | 公開日 | 2024-07-10 | | 実験手法 | X-RAY DIFFRACTION (1.92 Å) | | 主引用文献 | Uncommon P1 Anchor-featured Viral T Cell Epitope Preference within HLA-A*2601 and HLA-A*0101 Individuals.
Immunohorizons, 8, 2024
|
|
8XFZ
 
 | | The structure of HLA-A/L1-2 | | 分子名称: | Beta-2-microglobulin, HLA class I heavy chain, Major capsid protein L1 | | 著者 | Zhang, J.N, Yue, C, Liu, J. | | 登録日 | 2023-12-14 | | 公開日 | 2024-07-10 | | 実験手法 | X-RAY DIFFRACTION (2.32 Å) | | 主引用文献 | Uncommon P1 Anchor-featured Viral T Cell Epitope Preference within HLA-A*2601 and HLA-A*0101 Individuals.
Immunohorizons, 8, 2024
|
|
8XKC
 
 | | The structure of HLA-A/Pep16 | | 分子名称: | Beta-2-microglobulin, HLA class I heavy chain, Spike protein S1 | | 著者 | Zhang, J.N, Yue, C, Liu, J. | | 登録日 | 2023-12-23 | | 公開日 | 2024-07-10 | | 実験手法 | X-RAY DIFFRACTION (2.18 Å) | | 主引用文献 | Uncommon P1 Anchor-featured Viral T Cell Epitope Preference within HLA-A*2601 and HLA-A*0101 Individuals.
Immunohorizons, 8, 2024
|
|
6M1V
 
 | |
5JQ9
 
 | | Yersinia pestis DHPS with pterine-sulfa conjugate Compound 16 | | 分子名称: | 1,2-ETHANEDIOL, 4-{[(2-amino-4-oxo-3,4-dihydropteridin-6-yl)methyl]amino}-N-(3,4-dimethyl-1,2-oxazol-5-yl)benzene-1-sulfonamide, Dihydropteroate synthase | | 著者 | Wu, Y, White, S.W. | | 登録日 | 2016-05-04 | | 公開日 | 2016-08-10 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.101 Å) | | 主引用文献 | Pterin-sulfa conjugates as dihydropteroate synthase inhibitors and antibacterial agents.
Bioorg.Med.Chem.Lett., 26, 2016
|
|
5HGA
 
 | | HLA*A2402 complex with HIV nef138 Y2F-8mer mutant epitope | | 分子名称: | 8-mer from Protein Nef, Beta-2-microglobulin, HLA class I histocompatibility antigen, ... | | 著者 | Shi, Y, Qi, J, Gao, G.F. | | 登録日 | 2016-01-08 | | 公開日 | 2016-06-08 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.199 Å) | | 主引用文献 | Effects of a Single Escape Mutation on T Cell and HIV-1 Co-adaptation.
Cell Rep, 15, 2016
|
|
7WWV
 
 | | DNA bound-ICP1 Csy complex | | 分子名称: | Csy1, Csy2, Csy3, ... | | 著者 | Zhang, M, Peng, R. | | 登録日 | 2022-02-14 | | 公開日 | 2023-04-26 | | 最終更新日 | 2024-06-26 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Mechanistic insights into DNA binding and cleavage by a compact type I-F CRISPR-Cas system in bacteriophage.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
7WWU
 
 | | ICP1 Csy complex | | 分子名称: | Csy1, Csy2, Csy3, ... | | 著者 | Zhang, M, Peng, R. | | 登録日 | 2022-02-14 | | 公開日 | 2023-04-26 | | 最終更新日 | 2024-06-26 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | Mechanistic insights into DNA binding and cleavage by a compact type I-F CRISPR-Cas system in bacteriophage.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
7WKO
 
 | | ICP1 Csy1-2 complex | | 分子名称: | Csy1, Csy2 | | 著者 | Zhang, M, Peng, R. | | 登録日 | 2022-01-10 | | 公開日 | 2023-04-26 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Mechanistic insights into DNA binding and cleavage by a compact type I-F CRISPR-Cas system in bacteriophage.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
7WKP
 
 | | ICP1 Csy4 | | 分子名称: | 3' stem loop crRNA, Csy4 | | 著者 | Zhang, M, Peng, R. | | 登録日 | 2022-01-10 | | 公開日 | 2023-04-26 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Mechanistic insights into DNA binding and cleavage by a compact type I-F CRISPR-Cas system in bacteriophage.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8Y0Y
 
 | | Cryo-EM structure of the 123-316 scDb/PT-RBD complex | | 分子名称: | 123-316 scDb, 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | 著者 | Jia, G.W, Tong, Z, Tong, J.Y, Su, Z.M. | | 登録日 | 2024-01-23 | | 公開日 | 2024-07-17 | | 実験手法 | ELECTRON MICROSCOPY (2.86 Å) | | 主引用文献 | Deciphering a reliable synergistic bispecific strategy of rescuing antibodies for SARS-CoV-2 escape variants, including BA.2.86, EG.5.1, and JN.1.
Cell Rep, 43, 2024
|
|
8YGH
 
 | | pP1192R-apo open state | | 分子名称: | DNA topoisomerase 2 | | 著者 | Sun, J.Q, Liu, R.L. | | 登録日 | 2024-02-26 | | 公開日 | 2024-09-18 | | 実験手法 | ELECTRON MICROSCOPY (2.77 Å) | | 主引用文献 | Structural basis for difunctional mechanism of m-AMSA against African swine fever virus pP1192R.
Nucleic Acids Res., 2024
|
|
8YGE
 
 | |
8YGG
 
 | | pP1192R-apo Closed state | | 分子名称: | DNA topoisomerase 2 | | 著者 | Sun, J.Q, Liu, R.L. | | 登録日 | 2024-02-26 | | 公開日 | 2024-09-18 | | 実験手法 | ELECTRON MICROSCOPY (2.98 Å) | | 主引用文献 | Structural basis for difunctional mechanism of m-AMSA against African swine fever virus pP1192R.
Nucleic Acids Res., 2024
|
|
