6LBA
 
 | | Cryo-EM structure of the AtMLKL2 tetramer | | 分子名称: | Protein kinase family protein | | 著者 | Lisa, M, Huang, M, Zhang, X, Ryohei, T.N, Leila, B.K, Isabel, M.L.S, Florence, J, Viera, K, Dmitry, L, Jane, E.P, James, M.M, Kay, H, Paul, S.L, Chai, J, Takaki, M. | | 登録日 | 2019-11-13 | | 公開日 | 2020-11-18 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (4.1 Å) | | 主引用文献 | Cryo-EM structure of the AtMLKL3 tetramer
To Be Published
|
|
7DWQ
 
 | | Photosystem I from a chlorophyll d-containing cyanobacterium Acaryochloris marina | | 分子名称: | (6'R,11cis,11'cis,13cis,15cis)-4',5'-didehydro-5',6'-dihydro-beta,beta-carotene, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | 著者 | Chen, J.H, Zhang, X, Shen, J.R. | | 登録日 | 2021-01-17 | | 公開日 | 2021-06-02 | | 最終更新日 | 2025-07-02 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | A unique photosystem I reaction center from a chlorophyll d-containing cyanobacterium Acaryochloris marina.
J Integr Plant Biol, 63, 2021
|
|
6U6V
 
 | | Crystal structure of human PD-1H / VISTA | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, V-type immunoglobulin domain-containing suppressor of T-cell activation | | 著者 | Slater, B.T, Han, X, Chen, L, Xiong, Y. | | 登録日 | 2019-08-30 | | 公開日 | 2020-01-01 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structural insight into T cell coinhibition by PD-1H (VISTA).
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
7YE6
 
 | | BAM-EspP complex structure with BamA-N427C/EspP-R1297C mutations in nanodisc | | 分子名称: | Outer membrane protein assembly factor BamA, Outer membrane protein assembly factor BamB, Outer membrane protein assembly factor BamC, ... | | 著者 | Shen, C, Chang, S, Luo, Q, Zhang, Z, Luo, B, Lu, G, Zhu, X, Wei, X, Dong, C, Zhang, X, Tang, X, Dong, H. | | 登録日 | 2022-07-05 | | 公開日 | 2023-07-12 | | 最終更新日 | 2025-07-02 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Structural basis of BAM-mediated outer membrane beta-barrel protein assembly.
Nature, 617, 2023
|
|
7YE4
 
 | | BAM-EspP complex structure with BamA-G431C and G781C/EspP-N1293C and A1043C mutations in nanodisc | | 分子名称: | Outer membrane protein assembly factor BamA, Outer membrane protein assembly factor BamB, Outer membrane protein assembly factor BamC, ... | | 著者 | Shen, C, Chang, S, Luo, Q, Zhang, Z, Luo, B, Lu, G, Zhu, X, Wei, X, Dong, C, Zhang, X, Tang, X, Dong, H. | | 登録日 | 2022-07-05 | | 公開日 | 2023-07-12 | | 最終更新日 | 2025-06-25 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Structural basis of BAM-mediated outer membrane beta-barrel protein assembly.
Nature, 617, 2023
|
|
3WUB
 
 | | The wild type crystal structure of b-1,4-Xylanase (XynAS9) from Streptomyces sp. 9 | | 分子名称: | Endo-1,4-beta-xylanase A, ZINC ION | | 著者 | Chen, C.C, Han, X, Lv, P, Ko, T.P, Peng, W, Huang, C.H, Zheng, Y, Gao, J, Yang, Y.Y, Guo, R.T. | | 登録日 | 2014-04-23 | | 公開日 | 2014-10-29 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (2.08 Å) | | 主引用文献 | Structural perspectives of an engineered beta-1,4-xylanase with enhanced thermostability.
J.Biotechnol., 189C, 2014
|
|
3WUG
 
 | | The mutant crystal structure of b-1,4-Xylanase (XynAS9_V43P/G44E) with xylobiose from Streptomyces sp. 9 | | 分子名称: | Endo-1,4-beta-xylanase A, ZINC ION, beta-D-xylopyranose-(1-4)-beta-D-xylopyranose | | 著者 | Chen, C.C, Han, X, Lv, P, Ko, T.P, Peng, W, Huang, C.H, Zheng, Y, Gao, J, Yang, Y.Y, Guo, R.T. | | 登録日 | 2014-04-23 | | 公開日 | 2014-10-29 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (1.88 Å) | | 主引用文献 | Structural perspectives of an engineered beta-1,4-xylanase with enhanced thermostability.
J.Biotechnol., 189C, 2014
|
|
3WUE
 
 | | The wild type crystal structure of b-1,4-Xylanase (XynAS9) with xylobiose from Streptomyces sp. 9 | | 分子名称: | Endo-1,4-beta-xylanase A, ZINC ION, beta-D-xylopyranose-(1-4)-beta-D-xylopyranose | | 著者 | Chen, C.C, Han, X, Lv, P, Ko, T.P, Peng, W, Huang, C.H, Zheng, Y, Gao, J, Yang, Y, Guo, R.T. | | 登録日 | 2014-04-23 | | 公開日 | 2014-10-29 | | 最終更新日 | 2024-11-13 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | Structural perspectives of an engineered beta-1,4-xylanase with enhanced thermostability.
J.Biotechnol., 189C, 2014
|
|
3WUF
 
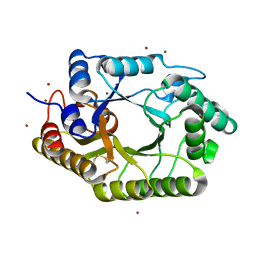 | | The mutant crystal structure of b-1,4-Xylanase (XynAS9_V43P/G44E) from Streptomyces sp. 9 | | 分子名称: | Endo-1,4-beta-xylanase A, ZINC ION | | 著者 | Chen, C.C, Han, X, Lv, P, Ko, T.P, Peng, W, Huang, C.H, Zheng, Y, Gao, J, Yang, Y.Y, Guo, R.T. | | 登録日 | 2014-04-23 | | 公開日 | 2014-10-29 | | 最終更新日 | 2024-11-13 | | 実験手法 | X-RAY DIFFRACTION (2.04 Å) | | 主引用文献 | Structural perspectives of an engineered beta-1,4-xylanase with enhanced thermostability.
J.Biotechnol., 189C, 2014
|
|
5M00
 
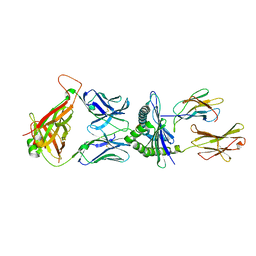 | | Crystal structure of murine P14 TCR complex with H-2Db and Y4A, modified gp33 peptide from LCMV | | 分子名称: | Beta-2-microglobulin, H-2 class I histocompatibility antigen, D-B alpha chain, ... | | 著者 | Achour, A, Sandalova, T, Sun, R, Han, X. | | 登録日 | 2016-10-03 | | 公開日 | 2017-12-20 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Thernary complexes of TCR P14 give insights into the mechanisms behind reestablishment of CTL responses against a viral escape mutant
To Be Published
|
|
5M01
 
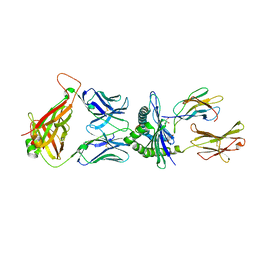 | | Crystal structure of murine P14 TCR/ H-2Db complex with PA, modified gp33 peptide from LCMV | | 分子名称: | Beta-2-microglobulin, GLYCEROL, H-2 class I histocompatibility antigen, ... | | 著者 | Achour, A, Sandalova, T, Sun, R, Han, X. | | 登録日 | 2016-10-03 | | 公開日 | 2017-10-25 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Thernary complexes of TCR P14 give insights into the mechanisms behind reestablishment of CTL responses against a viral escape mutant
To Be Published
|
|
7ARI
 
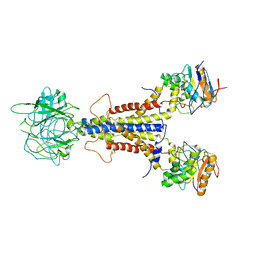 | | LolCDE apo structure | | 分子名称: | Lipoprotein-releasing ABC transporter permease subunit LolC, Lipoprotein-releasing system ATP-binding protein LolD, Lipoprotein-releasing system transmembrane protein LolE | | 著者 | Tang, X.D, Chang, S.H, Zhang, K, Wang, T, Luo, Q.H, Qiao, W, Wang, C, Zhang, Z.B, Zhang, Z.Y, Zhu, X.F, Dong, C.J, Zhang, X, Dong, H.H. | | 登録日 | 2020-10-25 | | 公開日 | 2021-04-07 | | 最終更新日 | 2025-07-02 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Structural basis for bacterial lipoprotein relocation by the transporter LolCDE.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7ARM
 
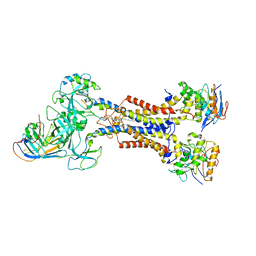 | | LolCDE in complex with lipoprotein and LolA | | 分子名称: | (2S)-3-hydroxypropane-1,2-diyl dihexadecanoate, LPP, Lipoprotein-releasing ABC transporter permease subunit LolC, ... | | 著者 | Tang, X.D, Chang, S.H, Zhang, K, Wang, T, Luo, Q.H, Qiao, W, Wang, C, Shen, C.R, Zhang, Z.B, Zhang, Z.Y, Zhu, X.F, Wei, X.W, Dong, C.J, Zhang, X, Dong, H.H. | | 登録日 | 2020-10-25 | | 公開日 | 2021-04-07 | | 最終更新日 | 2024-11-13 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Structural basis for bacterial lipoprotein relocation by the transporter LolCDE.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7ARL
 
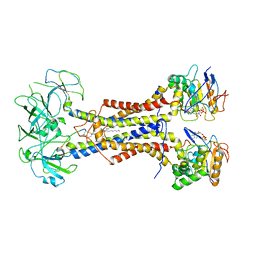 | | LolCDE in complex with lipoprotein and ADP | | 分子名称: | (2S)-3-hydroxypropane-1,2-diyl dihexadecanoate, ADENOSINE-5'-DIPHOSPHATE, LPP, ... | | 著者 | Tang, X.D, Chang, S.H, Zhang, K, Wang, T, Luo, Q.H, Qiao, W, Wang, C, Shen, C.R, Zhang, Z.B, Zhang, Z.Y, Zhu, X.F, Wei, X.W, Dong, C.J, Zhang, X, Dong, H.H. | | 登録日 | 2020-10-25 | | 公開日 | 2021-04-07 | | 最終更新日 | 2024-10-09 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Structural basis for bacterial lipoprotein relocation by the transporter LolCDE.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7ARK
 
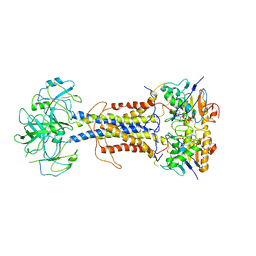 | | LolCDE in complex with AMP-PNP in the closed NBD state | | 分子名称: | Lipoprotein-releasing ABC transporter permease subunit LolC, Lipoprotein-releasing system ATP-binding protein LolD, Lipoprotein-releasing system transmembrane protein LolE, ... | | 著者 | Tang, X.D, Chang, S.H, Zhang, K, Wang, T, Luo, Q.H, Qiao, W, Wang, C, Zhang, Z.B, Zhang, Z.Y, Zhu, X.F, Dong, C.J, Zhang, X, Dong, H.H. | | 登録日 | 2020-10-25 | | 公開日 | 2021-04-07 | | 最終更新日 | 2024-05-01 | | 実験手法 | ELECTRON MICROSCOPY (4.1 Å) | | 主引用文献 | Structural basis for bacterial lipoprotein relocation by the transporter LolCDE.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7ARH
 
 | | LolCDE in complex with lipoprotein | | 分子名称: | (2S)-3-hydroxypropane-1,2-diyl dihexadecanoate, LPP, Lipoprotein-releasing ABC transporter permease subunit LolC, ... | | 著者 | Tang, X.D, Chang, S.H, Zhang, K, Wang, T, Luo, Q.H, Qiao, W, Wang, C, Zhang, Z.B, Zhang, Z.Y, Zhu, X.F, Dong, C.J, Zhang, X, Dong, H.H. | | 登録日 | 2020-10-25 | | 公開日 | 2021-04-07 | | 最終更新日 | 2025-07-09 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Structural basis for bacterial lipoprotein relocation by the transporter LolCDE.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7ARJ
 
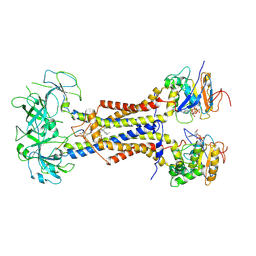 | | LolCDE in complex with lipoprotein and AMPPNP complex undimerized form | | 分子名称: | (2S)-3-hydroxypropane-1,2-diyl dihexadecanoate, LPP, Lipoprotein-releasing ABC transporter permease subunit LolC, ... | | 著者 | Tang, X.D, Chang, S.H, Zhang, K, Wang, T, Luo, Q.H, Qiao, W, Wang, C, Zhang, Z.B, Zhang, Z.Y, Zhu, X.F, Dong, C.J, Zhang, X, Dong, H.H. | | 登録日 | 2020-10-25 | | 公開日 | 2021-04-07 | | 最終更新日 | 2024-10-23 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Structural basis for bacterial lipoprotein relocation by the transporter LolCDE.
Nat.Struct.Mol.Biol., 28, 2021
|
|
8QN4
 
 | | Structure of BAM-EspP complex in the non-closing EspP state | | 分子名称: | EspP epsilon, Outer membrane protein assembly factor BamA, Outer membrane protein assembly factor BamB, ... | | 著者 | Xie, T, Pang, J, Shen, C, Chang, S, Tang, X, Zhang, X, Dong, H, Zhou, R. | | 登録日 | 2023-09-25 | | 公開日 | 2024-10-02 | | 最終更新日 | 2024-10-23 | | 実験手法 | ELECTRON MICROSCOPY (3.36 Å) | | 主引用文献 | Dynamic topology-mediated maturation of beta-barrel proteins in BAM-catalyzed folding
To Be Published
|
|
9IK1
 
 | | Cryo-EM structure of the human P2X3 receptor-compound 26a complex | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-[2-cyclopropyl-7-[[(1~{R})-1-naphthalen-2-ylethyl]amino]-[1,2,4]triazolo[1,5-a]pyrimidin-5-yl]piperazine-1-carboxamide, ADENOSINE-5'-TRIPHOSPHATE, ... | | 著者 | Kim, S, Kim, G.R, Kim, Y.O, Han, X, Nagel, J, Kim, J, Song, D.I, Muller, C.E, Yoon, M.H, Jin, M.S, Kim, Y.C. | | 登録日 | 2024-06-26 | | 公開日 | 2024-09-04 | | 最終更新日 | 2025-07-02 | | 実験手法 | ELECTRON MICROSCOPY (2.61 Å) | | 主引用文献 | Discovery of Triazolopyrimidine Derivatives as Selective P2X3 Receptor Antagonists Binding to an Unprecedented Allosteric Site as Evidenced by Cryo-Electron Microscopy.
J.Med.Chem., 67, 2024
|
|
1NP5
 
 | | (GAC)3 parallel duplex | | 分子名称: | 5'-D(*GP*AP*CP*GP*AP*CP*GP*AP*C)-3' | | 著者 | Zheng, M, Han, X, Gao, X. | | 登録日 | 2003-01-17 | | 公開日 | 2003-02-11 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Strand polarity of trinucleotide repeat sequences:
NMR studies of parallel/anti-pararell duplex,{d(GAC)3}2
To be Published
|
|
7XIL
 
 | | SARS-CoV-2-Beta-RBD and B38-GWP/P-VK antibody complex | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, B38 Fab heavy chain, B38 Fab light chain, ... | | 著者 | Shi, R, Wang, Y, Wu, Z, Han, X, Yan, J. | | 登録日 | 2022-04-13 | | 公開日 | 2023-04-26 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (2.91 Å) | | 主引用文献 | SARS-CoV-2-Beta-RBD and B38-GWP/P-VK antibody complex
To Be Published
|
|
7XIK
 
 | | SARS-CoV-2-Omicron-RBD and B38-GWP/P-VK antibody complex | | 分子名称: | B38 Fab heavy chain, B38 Fab light chain, Spike protein S1 | | 著者 | Shi, R, Wang, Y, Wu, Z, Han, X, Yan, J. | | 登録日 | 2022-04-13 | | 公開日 | 2023-04-26 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (2.89 Å) | | 主引用文献 | SARS-CoV-2-Omicron-RBD and B38-GWP/P-VK antibody complex
To Be Published
|
|
8IJT
 
 | | crystal structure of Hyp N135A mutant from Hypoxylon sp. E7406B | | 分子名称: | Terpene synthase | | 著者 | Gao, J, Su, L.Q, Li, Q, Han, X, Wei, H.L, Dai, Z.J, Liu, W.D. | | 登録日 | 2023-02-28 | | 公開日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.56 Å) | | 主引用文献 | crystal structure of Hyp N135A mutant from Hypoxylon sp. E7406B
to be published
|
|
8II9
 
 | | crystal structure of Hyp mutant from Hypoxylon sp. E7406B | | 分子名称: | Terpene synthase | | 著者 | Gao, J, Liu, W.D, Li, Q, Han, X, Wei, H.L, Dai, Z.J, Su, L.Q. | | 登録日 | 2023-02-24 | | 公開日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.17 Å) | | 主引用文献 | crystal structure of Hyp
to be published
|
|
8XTC
 
 | | Crystal structure of a novel PU plastic degradation urethanase UMG-SP2 mutant from uncultured bacterium in complex with ligand | | 分子名称: | 4-oxidanylbutyl ~{N}-(4-aminophenyl)carbamate, GLYCEROL, umgsp2-mut | | 著者 | Cong, L, Li, Z.S, Zheng, Z.R, Han, X, Gert, W, Wei, R, Liu, W.D, Bornscheuer, U.T. | | 登録日 | 2024-01-10 | | 公開日 | 2025-01-15 | | 最終更新日 | 2025-04-30 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Structure-Guided Engineering of a Versatile Urethanase Improves Its Polyurethane Depolymerization Activity.
Adv Sci, 12, 2025
|
|
