1S1V
 
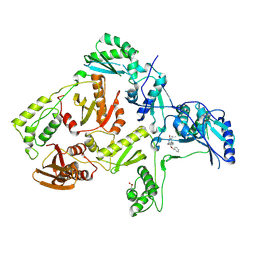 | | Crystal structure of L100I mutant HIV-1 reverse transcriptase in complex with TNK-651 | | 分子名称: | 6-BENZYL-1-BENZYLOXYMETHYL-5-ISOPROPYL URACIL, Reverse transcriptase | | 著者 | Ren, J, Nichols, C.E, Chamberlain, P.P, Stammers, D.K. | | 登録日 | 2004-01-07 | | 公開日 | 2004-06-29 | | 最終更新日 | 2021-11-10 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Crystal structures of HIV-1 reverse transcriptases mutated at codons 100, 106 and 108 and mechanisms of resistance to non-nucleoside inhibitors
J.Mol.Biol., 336, 2004
|
|
1S1U
 
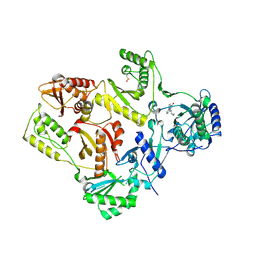 | | Crystal structure of L100I mutant HIV-1 reverse transcriptase in complex with nevirapine | | 分子名称: | 11-CYCLOPROPYL-5,11-DIHYDRO-4-METHYL-6H-DIPYRIDO[3,2-B:2',3'-E][1,4]DIAZEPIN-6-ONE, Reverse transcriptase | | 著者 | Ren, J, Nichols, C.E, Chamberlain, P.P, Stammers, D.K. | | 登録日 | 2004-01-07 | | 公開日 | 2004-06-29 | | 最終更新日 | 2021-11-10 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Crystal structures of HIV-1 reverse transcriptases mutated at codons 100, 106 and 108 and mechanisms of resistance to non-nucleoside inhibitors
J.Mol.Biol., 336, 2004
|
|
1S1W
 
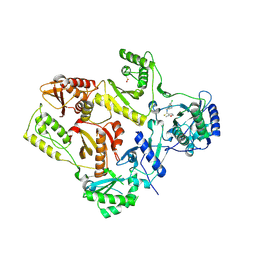 | | Crystal structure of V106A mutant HIV-1 reverse transcriptase in complex with UC-781 | | 分子名称: | 2-METHYL-FURAN-3-CARBOTHIOIC ACID [4-CHLORO-3-(3-METHYL-BUT-2-ENYLOXY)-PHENYL]-AMIDE, Reverse transcriptase | | 著者 | Ren, J, Nichols, C.E, Chamberlain, P.P, Stammers, D.K. | | 登録日 | 2004-01-07 | | 公開日 | 2004-06-29 | | 最終更新日 | 2021-11-10 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Crystal structures of HIV-1 reverse transcriptases mutated at codons 100, 106 and 108 and mechanisms of resistance to non-nucleoside inhibitors
J.Mol.Biol., 336, 2004
|
|
9GPB
 
 | |
7GPB
 
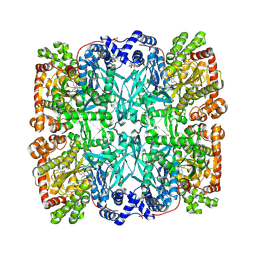 | | STRUCTURAL MECHANISM FOR GLYCOGEN PHOSPHORYLASE CONTROL BY PHOSPHORYLATION AND AMP | | 分子名称: | ADENOSINE MONOPHOSPHATE, GLYCOGEN PHOSPHORYLASE B, PYRIDOXAL-5'-PHOSPHATE, ... | | 著者 | Barford, D, Hu, S.-H, Johnson, L.N. | | 登録日 | 1990-11-13 | | 公開日 | 1992-10-15 | | 最終更新日 | 2017-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Structural mechanism for glycogen phosphorylase control by phosphorylation and AMP.
J.Mol.Biol., 218, 1991
|
|
7NXA
 
 | | Crystal structure of the receptor binding domain of SARS-CoV-2 B.1.351 variant Spike glycoprotein in complex with COVOX-222 and EY6A Fabs | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, COVOX-222 Fab heavy chain, COVOX-222 Fab light chain, ... | | 著者 | Zhou, D, Ren, J, Stuart, D. | | 登録日 | 2021-03-17 | | 公開日 | 2021-04-07 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Antibody evasion by the P.1 strain of SARS-CoV-2.
Cell, 184, 2021
|
|
7NXB
 
 | | Crystal structure of the receptor binding domain of SARS-CoV-2 P.1 variant Spike glycoprotein in complex with COVOX-222 and EY6A Fabs | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, COVOX-222 Fab heavy chain, COVOX-222 Fab light chain, ... | | 著者 | Zhou, D, Ren, J, Stuart, D. | | 登録日 | 2021-03-17 | | 公開日 | 2021-04-07 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.67 Å) | | 主引用文献 | Antibody evasion by the P.1 strain of SARS-CoV-2.
Cell, 184, 2021
|
|
7NX6
 
 | | Crystal structure of the receptor binding domain of SARS-CoV-2 Spike glycoprotein in complex with COVOX-222 and EY6A Fabs | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, COVOX-222 Fab Heavy chain, ... | | 著者 | Zhou, D, Ren, J, Stuart, D. | | 登録日 | 2021-03-17 | | 公開日 | 2021-04-07 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Antibody evasion by the P.1 strain of SARS-CoV-2.
Cell, 184, 2021
|
|
7NXC
 
 | |
7NX7
 
 | | Crystal structure of the K417N mutant receptor binding domain of SARS-CoV-2 Spike glycoprotein in complex with COVOX-222 and EY6A Fabs | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, CITRIC ACID, ... | | 著者 | Zhou, D, Ren, J, Stuart, D. | | 登録日 | 2021-03-17 | | 公開日 | 2021-04-07 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Antibody evasion by the P.1 strain of SARS-CoV-2.
Cell, 184, 2021
|
|
7NX9
 
 | | Crystal structure of the N501Y mutant receptor binding domain of SARS-CoV-2 Spike glycoprotein in complex with COVOX-222 and EY6A Fabs | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, COVOX-222 Fab heavy chain, ... | | 著者 | Zhou, D, Ren, J, Stuart, D. | | 登録日 | 2021-03-17 | | 公開日 | 2021-04-07 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Antibody evasion by the P.1 strain of SARS-CoV-2.
Cell, 184, 2021
|
|
7NX8
 
 | | Crystal structure of the K417T mutant receptor binding domain of SARS-CoV-2 Spike glycoprotein in complex with COVOX-222 and EY6A Fabs | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, CITRIC ACID, ... | | 著者 | Zhou, D, Ren, J, Stuart, D. | | 登録日 | 2021-03-17 | | 公開日 | 2021-04-07 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Antibody evasion by the P.1 strain of SARS-CoV-2.
Cell, 184, 2021
|
|
5WSN
 
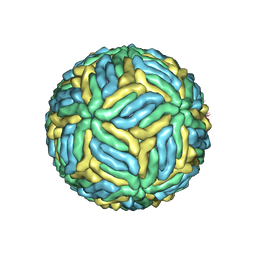 | | Structure of Japanese encephalitis virus | | 分子名称: | E protein, M protein | | 著者 | Wang, X, Zhu, L, Li, S, Yuan, S, Qin, C, Fry, E.E, Stuart, I.D, Rao, Z. | | 登録日 | 2016-12-07 | | 公開日 | 2017-05-17 | | 最終更新日 | 2019-11-06 | | 実験手法 | ELECTRON MICROSCOPY (4.3 Å) | | 主引用文献 | Near-atomic structure of Japanese encephalitis virus reveals critical determinants of virulence and stability
Nat Commun, 8, 2017
|
|
5NEU
 
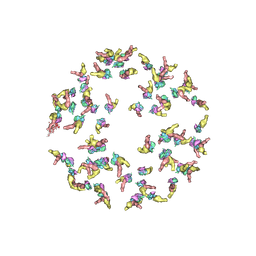 | |
5NE4
 
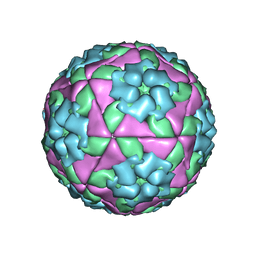 | |
5NED
 
 | |
4GPB
 
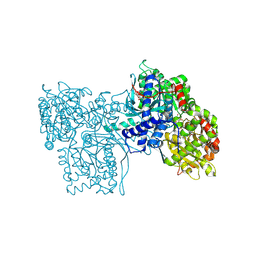 | |
3GPB
 
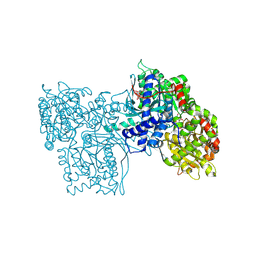 | |
5GPB
 
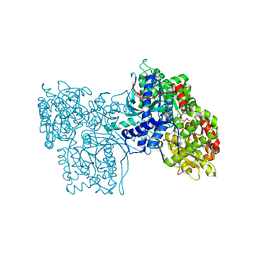 | |
1RT2
 
 | | CRYSTAL STRUCTURE OF HIV-1 REVERSE TRANSCRIPTASE COMPLEXED WITH TNK-651 | | 分子名称: | 6-BENZYL-1-BENZYLOXYMETHYL-5-ISOPROPYL URACIL, HIV-1 REVERSE TRANSCRIPTASE | | 著者 | Ren, J, Esnouf, R, Hopkins, A, Willcox, B, Jones, Y, Ross, C, Stammers, D, Stuart, D. | | 登録日 | 1996-03-16 | | 公開日 | 1997-04-21 | | 最終更新日 | 2023-08-09 | | 実験手法 | X-RAY DIFFRACTION (2.55 Å) | | 主引用文献 | Complexes of HIV-1 reverse transcriptase with inhibitors of the HEPT series reveal conformational changes relevant to the design of potent non-nucleoside inhibitors.
J.Med.Chem., 39, 1996
|
|
1RT1
 
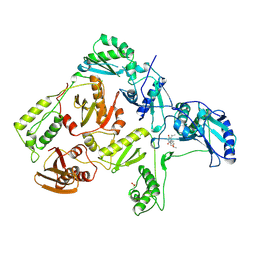 | | CRYSTAL STRUCTURE OF HIV-1 REVERSE TRANSCRIPTASE COMPLEXED WITH MKC-442 | | 分子名称: | 6-BENZYL-1-ETHOXYMETHYL-5-ISOPROPYL URACIL, HIV-1 REVERSE TRANSCRIPTASE | | 著者 | Ren, J, Esnouf, R, Hopkins, A, Willcox, B, Jones, Y, Ross, C, Stammers, D, Stuart, D. | | 登録日 | 1996-03-16 | | 公開日 | 1997-04-21 | | 最終更新日 | 2023-08-09 | | 実験手法 | X-RAY DIFFRACTION (2.55 Å) | | 主引用文献 | Complexes of HIV-1 reverse transcriptase with inhibitors of the HEPT series reveal conformational changes relevant to the design of potent non-nucleoside inhibitors.
J.Med.Chem., 39, 1996
|
|
1RTJ
 
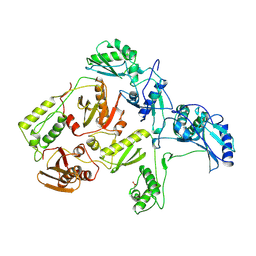 | | MECHANISM OF INHIBITION OF HIV-1 REVERSE TRANSCRIPTASE BY NON-NUCLEOSIDE INHIBITORS | | 分子名称: | HIV-1 REVERSE TRANSCRIPTASE | | 著者 | Ren, J, Esnouf, R, Ross, C, Jones, Y, Stammers, D, Stuart, D. | | 登録日 | 1995-05-03 | | 公開日 | 1996-04-03 | | 最終更新日 | 2024-06-05 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | Mechanism of inhibition of HIV-1 reverse transcriptase by non-nucleoside inhibitors.
Nat.Struct.Biol., 2, 1995
|
|
6XF8
 
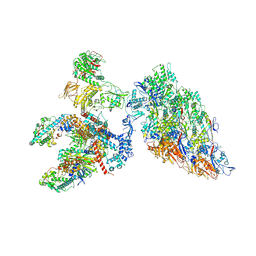 | | DLP 5 fold | | 分子名称: | Inner capsid protein lambda-1, Inner capsid protein sigma-2, Outer capsid protein mu-1, ... | | 著者 | Sutton, G, Sun, D.P, Fu, X.F, Kotecha, A, Hecksel, G.W, Clare, D.K, Zhang, P, Stuart, D, Boyce, M. | | 登録日 | 2020-06-15 | | 公開日 | 2020-09-23 | | 実験手法 | ELECTRON MICROSCOPY (6.5 Å) | | 主引用文献 | Assembly intermediates of orthoreovirus captured in the cell.
Nat Commun, 11, 2020
|
|
5NER
 
 | |
5NEJ
 
 | |
