7JYE
 
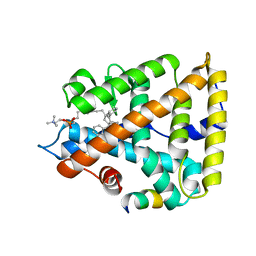 | | Human Liver Receptor Homolog-1 in Complex with 9ChoP and a Fragment of Tif2 | | 分子名称: | 9-[(3~{a}~{R},6~{R},6~{a}~{R})-6-oxidanyl-3-phenyl-3~{a}-(1-phenylethenyl)-4,5,6,6~{a}-tetrahydro-1~{H}-pentalen-2-yl]nonyl 2-(trimethyl-$l^{4}-azanyl)ethyl hydrogen phosphate, Nuclear receptor coactivator 2, Nuclear receptor subfamily 5 group A member 2 | | 著者 | D'Agostino, E.H, Mays, S.G, Ortlund, E.A. | | 登録日 | 2020-08-30 | | 公開日 | 2022-03-02 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.551 Å) | | 主引用文献 | Tapping into a phospholipid-LRH-1 axis yields a powerful anti-inflammatory agent with in vivo activity against colitis
To Be Published
|
|
7CYC
 
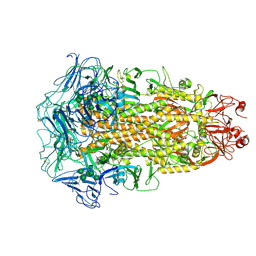 | | Cryo-EM structures of Alphacoronavirus spike glycoprotein | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | 著者 | Song, X, Shi, Y, Ding, W, Liu, Z.J, Peng, G. | | 登録日 | 2020-09-03 | | 公開日 | 2020-12-23 | | 最終更新日 | 2021-06-02 | | 実験手法 | ELECTRON MICROSCOPY (3.21 Å) | | 主引用文献 | Cryo-EM analysis of the HCoV-229E spike glycoprotein reveals dynamic prefusion conformational changes.
Nat Commun, 12, 2021
|
|
7CYD
 
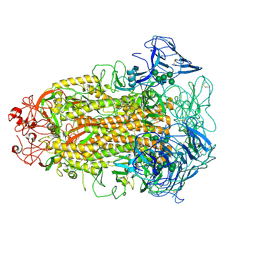 | | Cryo-EM structures of Alphacoronavirus spike glycoprotein | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-3)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Song, X, Shi, Y, Ding, W, Liu, Z.J, Peng, G. | | 登録日 | 2020-09-03 | | 公開日 | 2020-12-23 | | 最終更新日 | 2021-06-02 | | 実験手法 | ELECTRON MICROSCOPY (3.55 Å) | | 主引用文献 | Cryo-EM analysis of the HCoV-229E spike glycoprotein reveals dynamic prefusion conformational changes.
Nat Commun, 12, 2021
|
|
7EOT
 
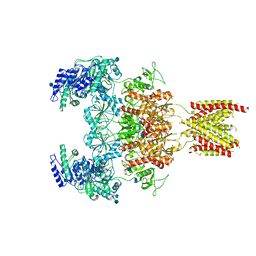 | |
7EOQ
 
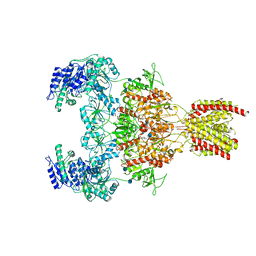 | |
7EOU
 
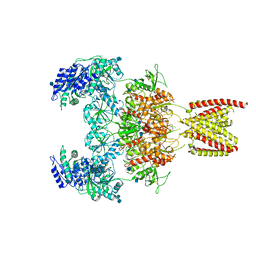 | | Structure of the human GluN1/GluN2A NMDA receptor in the glycine/glutamate/GNE-6901/9-AA bound state | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 7-[(4-fluoranylphenoxy)methyl]-3-[(1~{R},2~{R})-2-(hydroxymethyl)cyclopropyl]-2-methyl-[1,3]thiazolo[3,2-a]pyrimidin-5-one, 9-AMINOACRIDINE, ... | | 著者 | Wang, H, Zhu, S. | | 登録日 | 2021-04-22 | | 公開日 | 2021-06-30 | | 最終更新日 | 2021-08-18 | | 実験手法 | ELECTRON MICROSCOPY (4.3 Å) | | 主引用文献 | Gating mechanism and a modulatory niche of human GluN1-GluN2A NMDA receptors.
Neuron, 109, 2021
|
|
7EOS
 
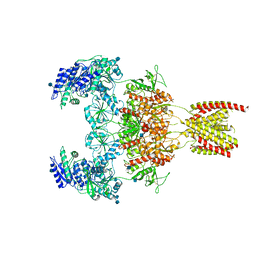 | |
7EOR
 
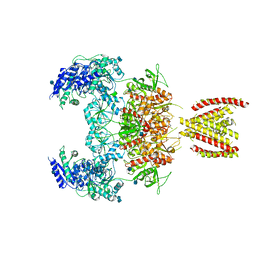 | | Structure of the human GluN1/GluN2A NMDA receptor in the glycine/glutamate/GNE-6901 bound state | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 7-[(4-fluoranylphenoxy)methyl]-3-[(1~{R},2~{R})-2-(hydroxymethyl)cyclopropyl]-2-methyl-[1,3]thiazolo[3,2-a]pyrimidin-5-one, Glutamate receptor ionotropic, ... | | 著者 | Wang, H, Zhu, S. | | 登録日 | 2021-04-22 | | 公開日 | 2021-06-30 | | 最終更新日 | 2021-08-18 | | 実験手法 | ELECTRON MICROSCOPY (4 Å) | | 主引用文献 | Gating mechanism and a modulatory niche of human GluN1-GluN2A NMDA receptors.
Neuron, 109, 2021
|
|
7DNO
 
 | | Characterization of Peptide Ligands Against WDR5 Isolated Using Phage Display Technique | | 分子名称: | CYS-ARG-THR-LEU-PRO-PHE, WD repeat-containing protein 5 | | 著者 | Cao, J, Cao, D, Xiong, B, Li, Y, Fan, T. | | 登録日 | 2020-12-10 | | 公開日 | 2021-02-10 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.03 Å) | | 主引用文献 | Phage-Display Based Discovery and Characterization of Peptide Ligands against WDR5.
Molecules, 26, 2021
|
|
7DC3
 
 | | Crystal structure of the MyRF ICA domain | | 分子名称: | Isoform 2 of Myelin regulatory factor | | 著者 | Shi, N, Wu, P, Zhen, X.K, Li, B.W. | | 登録日 | 2020-10-23 | | 公開日 | 2021-08-25 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Crystal structure of the MyRF ICA domain with its upstream beta-helical stalk reveals the molecular mechanisms underlying its trimerization and self-cleavage.
Int J Biol Sci, 17, 2021
|
|
7X0X
 
 | |
7X0Y
 
 | |
7UB0
 
 | |
7UB6
 
 | |
7UB5
 
 | |
7WH9
 
 | | holo structure of emodin 1-OH O-methyltransferase complex with emodin and S-Adenosyl-L-homocysteine | | 分子名称: | 3-METHYL-1,6,8-TRIHYDROXYANTHRAQUINONE, O-methyltransferase gedA, S-ADENOSYL-L-HOMOCYSTEINE | | 著者 | Liang, Y.J, Lu, X.F, Qi, F.F, Xue, Y.Y. | | 登録日 | 2021-12-30 | | 公開日 | 2023-01-11 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (2.803 Å) | | 主引用文献 | Characterization and Structural Analysis of Emodin- O -Methyltransferase from Aspergillus terreus.
J.Agric.Food Chem., 70, 2022
|
|
7EN0
 
 | | Structure and Activity of SLAC1 Channels for Stomatal Signaling in Leaves | | 分子名称: | DIUNDECYL PHOSPHATIDYL CHOLINE, SLow Anion Channel 1, SPHINGOSINE | | 著者 | Deng, Y, Kashtoh, H, Wang, Q, Zhen, G, Li, Q, Tang, L, Gao, H, Zhang, C, Qin, L, Su, M, Li, F, Huang, X, Wang, Y, Xie, Q, Clarke, O.B, Hendrickson, W.A, Chen, Y. | | 登録日 | 2021-04-15 | | 公開日 | 2021-05-19 | | 最終更新日 | 2024-06-05 | | 実験手法 | ELECTRON MICROSCOPY (2.97 Å) | | 主引用文献 | Structure and activity of SLAC1 channels for stomatal signaling in leaves.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
8I7O
 
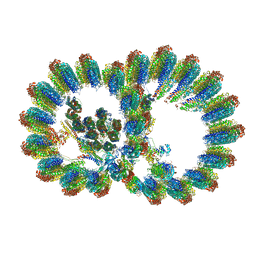 | | In situ structure of axonemal doublet microtubules in mouse sperm with 16-nm repeat | | 分子名称: | Cilia and flagella-associated protein 77, Cilia- and flagella-associated protein 20, Cilia- and flagella-associated protein 276, ... | | 著者 | Zhu, Y, Yin, G.L, Tai, L.H, Sun, F. | | 登録日 | 2023-02-01 | | 公開日 | 2023-10-11 | | 最終更新日 | 2023-12-06 | | 実験手法 | ELECTRON MICROSCOPY (4.5 Å) | | 主引用文献 | In-cell structural insight into the stability of sperm microtubule doublet.
Cell Discov, 9, 2023
|
|
8I7R
 
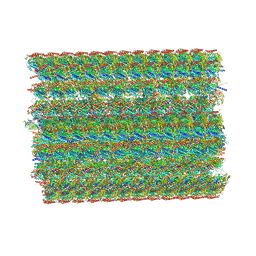 | | In situ structure of axonemal doublet microtubules in mouse sperm with 48-nm repeat | | 分子名称: | Cilia and flagella-associated protein 77, Cilia- and flagella-associated protein 107, Cilia- and flagella-associated protein 141, ... | | 著者 | Zhu, Y, Yin, G.L, Tai, L.H, Sun, F. | | 登録日 | 2023-02-02 | | 公開日 | 2023-10-11 | | 最終更新日 | 2023-12-06 | | 実験手法 | ELECTRON MICROSCOPY (6.5 Å) | | 主引用文献 | In-cell structural insight into the stability of sperm microtubule doublet.
Cell Discov, 9, 2023
|
|
8IVM
 
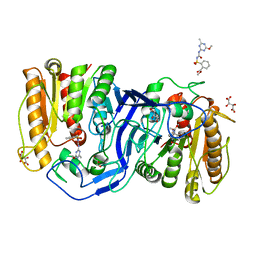 | | crystal structure of SulE mutant | | 分子名称: | Alpha/beta fold hydrolase, GLYCEROL, L(+)-TARTARIC ACID, ... | | 著者 | Liu, B, He, J, Ran, T, Wang, W. | | 登録日 | 2023-03-28 | | 公開日 | 2023-08-02 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (1.32 Å) | | 主引用文献 | Crystal structures of herbicide-detoxifying esterase reveal a lid loop affecting substrate binding and activity.
Nat Commun, 14, 2023
|
|
8IVS
 
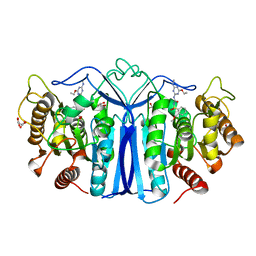 | | crystal structure of SulE mutant | | 分子名称: | Alpha/beta fold hydrolase, GLYCEROL, L(+)-TARTARIC ACID, ... | | 著者 | Liu, B, He, J, Ran, T, Wang, W. | | 登録日 | 2023-03-28 | | 公開日 | 2023-08-02 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (1.52 Å) | | 主引用文献 | Crystal structures of herbicide-detoxifying esterase reveal a lid loop affecting substrate binding and activity.
Nat Commun, 14, 2023
|
|
8J7J
 
 | | crystal structure of SulE mutant | | 分子名称: | Alpha/beta fold hydrolase, GLYCEROL, L(+)-TARTARIC ACID, ... | | 著者 | Liu, B, He, J, Ran, T, Wang, W. | | 登録日 | 2023-04-27 | | 公開日 | 2023-08-02 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (1.54 Å) | | 主引用文献 | Crystal structures of herbicide-detoxifying esterase reveal a lid loop affecting substrate binding and activity.
Nat Commun, 14, 2023
|
|
8IW6
 
 | | crystal structure of SulE mutant | | 分子名称: | Alpha/beta fold hydrolase, GLYCEROL, L(+)-TARTARIC ACID, ... | | 著者 | Liu, B, He, J, Ran, T, Wang, W. | | 登録日 | 2023-03-29 | | 公開日 | 2023-08-02 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (1.44 Å) | | 主引用文献 | Crystal structures of herbicide-detoxifying esterase reveal a lid loop affecting substrate binding and activity.
Nat Commun, 14, 2023
|
|
8IVE
 
 | | crystal structure of SulE mutant | | 分子名称: | 2-[[[[(4-CHLORO-6-METHOXY-2-PYRIMIDINYL)AMINO]CARBONYL]AMINO]SULFONYL]BENZOIC ACID ETHYL ESTER, Alpha/beta fold hydrolase, GLYCEROL | | 著者 | Liu, B, He, J, Ran, T, Wang, W. | | 登録日 | 2023-03-27 | | 公開日 | 2023-08-02 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (1.44 Å) | | 主引用文献 | Crystal structures of herbicide-detoxifying esterase reveal a lid loop affecting substrate binding and activity.
Nat Commun, 14, 2023
|
|
8IVN
 
 | | crystal structure of SulE mutant | | 分子名称: | Alpha/beta fold hydrolase, GLYCEROL, L(+)-TARTARIC ACID, ... | | 著者 | Liu, B, He, J, Ran, T, Wang, W. | | 登録日 | 2023-03-28 | | 公開日 | 2023-08-02 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Crystal structures of herbicide-detoxifying esterase reveal a lid loop affecting substrate binding and activity.
Nat Commun, 14, 2023
|
|
