5Z1V
 
 | | Crystal structure of AvrPib | | 分子名称: | AvrPib protein | | 著者 | Zhang, X, He, D, Zhao, Y.X, Taylor, I.A, Peng, Y.L, Yang, J, Liu, J.F. | | 登録日 | 2017-12-28 | | 公開日 | 2018-09-05 | | 最終更新日 | 2018-10-03 | | 実験手法 | X-RAY DIFFRACTION (1.661 Å) | | 主引用文献 | A positive-charged patch and stabilized hydrophobic core are essential for avirulence function of AvrPib in the rice blast fungus.
Plant J., 96, 2018
|
|
1F3L
 
 | |
1ORH
 
 | |
8H0Z
 
 | | Structure of SARS-CoV-1 Spike Protein with Engineered x1 Disulfide (S370C and D967C), Locked-122 Conformation | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BILIVERDINE IX ALPHA, ... | | 著者 | Zhang, X, Li, Z, Liu, Y, Wang, J, Fu, L, Wang, P, He, J, Xiong, X. | | 登録日 | 2022-09-30 | | 公開日 | 2022-11-09 | | 最終更新日 | 2023-07-19 | | 実験手法 | ELECTRON MICROSCOPY (2.99 Å) | | 主引用文献 | Disulfide stabilization reveals conserved dynamic features between SARS-CoV-1 and SARS-CoV-2 spikes.
Life Sci Alliance, 6, 2023
|
|
1PEG
 
 | | Structural basis for the product specificity of histone lysine methyltransferases | | 分子名称: | Histone H3, S-ADENOSYL-L-HOMOCYSTEINE, ZINC ION, ... | | 著者 | Zhang, X, Yang, Z, Khan, S.I, Horton, J.R, Tamaru, H, Selker, E.U, Cheng, X. | | 登録日 | 2003-05-21 | | 公開日 | 2003-08-05 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (2.59 Å) | | 主引用文献 | Structural basis for the product specificity of histone lysine methyltransferases
Mol.Cell, 12, 2003
|
|
1ORI
 
 | |
1OR8
 
 | |
1NCN
 
 | |
5YZG
 
 | | The Cryo-EM Structure of Human Catalytic Step I Spliceosome (C complex) at 4.1 angstrom resolution | | 分子名称: | 116 kDa U5 small nuclear ribonucleoprotein component, ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ... | | 著者 | Zhan, X, Yan, C, Zhang, X, Lei, J, Shi, Y. | | 登録日 | 2017-12-14 | | 公開日 | 2018-08-08 | | 最終更新日 | 2020-10-14 | | 実験手法 | ELECTRON MICROSCOPY (4.1 Å) | | 主引用文献 | Structure of a human catalytic step I spliceosome
Science, 359, 2018
|
|
5Z58
 
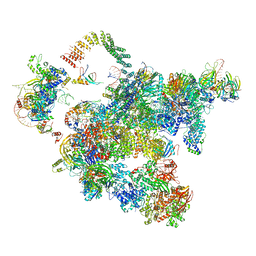 | | Cryo-EM structure of a human activated spliceosome (early Bact) at 4.9 angstrom. | | 分子名称: | 116 kDa U5 small nuclear ribonucleoprotein component, BUD13 homolog, Cell division cycle 5-like protein, ... | | 著者 | Zhang, X, Yan, C, Zhan, X, Li, L, Lei, J, Shi, Y. | | 登録日 | 2018-01-17 | | 公開日 | 2018-09-19 | | 最終更新日 | 2020-10-14 | | 実験手法 | ELECTRON MICROSCOPY (4.9 Å) | | 主引用文献 | Structure of the human activated spliceosome in three conformational states.
Cell Res., 28, 2018
|
|
5Z57
 
 | | Cryo-EM structure of the human activated spliceosome (late Bact) at 6.5 angstrom | | 分子名称: | 116 kDa U5 small nuclear ribonucleoprotein component, ALANINE, BUD13 homolog, ... | | 著者 | Zhang, X, Yan, C, Zhan, X, Li, L, Lei, J, Shi, Y. | | 登録日 | 2018-01-17 | | 公開日 | 2018-09-19 | | 最終更新日 | 2020-10-14 | | 実験手法 | ELECTRON MICROSCOPY (6.5 Å) | | 主引用文献 | Structure of the human activated spliceosome in three conformational states.
Cell Res., 28, 2018
|
|
5Z56
 
 | | cryo-EM structure of a human activated spliceosome (mature Bact) at 5.1 angstrom. | | 分子名称: | 116 kDa U5 small nuclear ribonucleoprotein component, BUD13 homolog, Cell division cycle 5-like protein, ... | | 著者 | Zhang, X, Yan, C, Zhan, X, Li, L, Lei, J, Shi, Y. | | 登録日 | 2018-01-17 | | 公開日 | 2018-09-19 | | 最終更新日 | 2020-10-14 | | 実験手法 | ELECTRON MICROSCOPY (5.1 Å) | | 主引用文献 | Structure of the human activated spliceosome in three conformational states.
Cell Res., 28, 2018
|
|
3AH5
 
 | | Crystal Structure of flavin dependent thymidylate synthase ThyX from helicobacter pylori complexed with FAD and dUMP | | 分子名称: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, FLAVIN-ADENINE DINUCLEOTIDE, SULFATE ION, ... | | 著者 | Zhang, X, Zhang, J, Hu, Y, Zou, Q, Wang, D. | | 登録日 | 2010-04-14 | | 公開日 | 2011-04-20 | | 最終更新日 | 2024-10-09 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Crystal structure and functional analysis of a flavin dependent thymidylate synthase from helicobacter pylori
To be Published
|
|
1H0C
 
 | | The crystal structure of human alanine:glyoxylate aminotransferase | | 分子名称: | (AMINOOXY)ACETIC ACID, GLYCEROL, PYRIDOXAL-5'-PHOSPHATE, ... | | 著者 | Zhang, X, Danpure, C.J, Roe, S.M, Pearl, L.H. | | 登録日 | 2002-06-17 | | 公開日 | 2003-06-12 | | 最終更新日 | 2019-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Crystal Structure of Alanine:Glyoxylate Aminotransferase and the Relationship between Genotype and Enzymatic Phenotype in Primary Hyperoxaluria Type 1.
J.Mol.Biol., 331, 2003
|
|
6J4A
 
 | |
6J4M
 
 | | Thermal treated soybean seed H-2 ferritin | | 分子名称: | Ferritin, MAGNESIUM ION | | 著者 | Zhang, X, Zang, J, Chen, H, Zhou, K, Zhao, G. | | 登録日 | 2019-01-09 | | 公開日 | 2019-09-18 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.598 Å) | | 主引用文献 | Thermostability of protein nanocages: the effect of natural extra peptide on the exterior surface.
Rsc Adv, 9, 2019
|
|
8H10
 
 | | Structure of SARS-CoV-1 Spike Protein with Engineered x1 Disulfide (S370C and D967C), Locked-2 Conformation | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BILIVERDINE IX ALPHA, ... | | 著者 | Zhang, X, Li, Z, Liu, Y, Wang, J, Fu, L, Wang, P, He, J, Xiong, X. | | 登録日 | 2022-09-30 | | 公開日 | 2022-10-19 | | 最終更新日 | 2023-07-19 | | 実験手法 | ELECTRON MICROSCOPY (2.99 Å) | | 主引用文献 | Disulfide stabilization reveals conserved dynamic features between SARS-CoV-1 and SARS-CoV-2 spikes.
Life Sci Alliance, 6, 2023
|
|
8H13
 
 | | Structure of SARS-CoV-1 Spike Protein with Engineered x2 Disulfide (G400C and V969C), Closed Conformation | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | 著者 | Zhang, X, Li, Z, Liu, Y, Wang, J, Fu, L, Wang, P, He, J, Xiong, X. | | 登録日 | 2022-09-30 | | 公開日 | 2022-10-19 | | 最終更新日 | 2023-07-19 | | 実験手法 | ELECTRON MICROSCOPY (4.05 Å) | | 主引用文献 | Disulfide stabilization reveals conserved dynamic features between SARS-CoV-1 and SARS-CoV-2 spikes.
Life Sci Alliance, 6, 2023
|
|
8H14
 
 | | Structure of SARS-CoV-1 Spike Protein with Engineered x3 Disulfide (D414C and V969C), Locked-1 Conformation | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, LINOLEIC ACID, Spike glycoprotein | | 著者 | Zhang, X, Li, Z, Liu, Y, Wang, J, Fu, L, Wang, P, He, J, Xiong, X. | | 登録日 | 2022-09-30 | | 公開日 | 2022-10-19 | | 最終更新日 | 2024-10-09 | | 実験手法 | ELECTRON MICROSCOPY (3.39 Å) | | 主引用文献 | Disulfide stabilization reveals conserved dynamic features between SARS-CoV-1 and SARS-CoV-2 spikes.
Life Sci Alliance, 6, 2023
|
|
8H16
 
 | | Structure of SARS-CoV-1 Spike Protein (S/native) at pH 5.5, Open Conformation | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | 著者 | Zhang, X, Li, Z, Liu, Y, Wang, J, Fu, L, Wang, P, He, J, Xiong, X. | | 登録日 | 2022-09-30 | | 公開日 | 2022-11-09 | | 最終更新日 | 2023-07-19 | | 実験手法 | ELECTRON MICROSCOPY (3.35534 Å) | | 主引用文献 | Disulfide stabilization reveals conserved dynamic features between SARS-CoV-1 and SARS-CoV-2 spikes.
Life Sci Alliance, 6, 2023
|
|
8H11
 
 | | Structure of SARS-CoV-1 Spike Protein with Engineered x1 Disulfide (S370C and D967C), Closed Conformation | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | 著者 | Zhang, X, Li, Z, Liu, Y, Wang, J, Fu, L, Wang, P, He, J, Xiong, X. | | 登録日 | 2022-09-30 | | 公開日 | 2022-11-09 | | 最終更新日 | 2023-07-19 | | 実験手法 | ELECTRON MICROSCOPY (2.72 Å) | | 主引用文献 | Disulfide stabilization reveals conserved dynamic features between SARS-CoV-1 and SARS-CoV-2 spikes.
Life Sci Alliance, 6, 2023
|
|
8H0Y
 
 | | Structure of SARS-CoV-1 Spike Protein with Engineered x1 Disulfide (S370C and D967C), Locked-112 Conformation | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, BILIVERDINE IX ALPHA, LINOLEIC ACID, ... | | 著者 | Zhang, X, Li, Z, Liu, Y, Wang, J, Fu, L, Wang, P, He, J, Xiong, X. | | 登録日 | 2022-09-30 | | 公開日 | 2022-11-09 | | 最終更新日 | 2023-07-19 | | 実験手法 | ELECTRON MICROSCOPY (2.85 Å) | | 主引用文献 | Disulfide stabilization reveals conserved dynamic features between SARS-CoV-1 and SARS-CoV-2 spikes.
Life Sci Alliance, 6, 2023
|
|
8H12
 
 | | Structure of SARS-CoV-1 Spike Protein with Engineered x2 Disulfide (G400C and V969C), Locked-2 Conformation | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | 著者 | Zhang, X, Li, Z, Liu, Y, Wang, J, Fu, L, Wang, P, He, J, Xiong, X. | | 登録日 | 2022-09-30 | | 公開日 | 2022-11-09 | | 最終更新日 | 2023-07-19 | | 実験手法 | ELECTRON MICROSCOPY (3.44681 Å) | | 主引用文献 | Disulfide stabilization reveals conserved dynamic features between SARS-CoV-1 and SARS-CoV-2 spikes.
Life Sci Alliance, 6, 2023
|
|
8H15
 
 | | Structure of SARS-CoV-1 Spike Protein (S/native) at pH 5.5, Closed Conformation | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | 著者 | Zhang, X, Li, Z, Liu, Y, Wang, J, Fu, L, Wang, P, He, J, Xiong, X. | | 登録日 | 2022-09-30 | | 公開日 | 2022-11-09 | | 最終更新日 | 2023-07-19 | | 実験手法 | ELECTRON MICROSCOPY (3.14182 Å) | | 主引用文献 | Disulfide stabilization reveals conserved dynamic features between SARS-CoV-1 and SARS-CoV-2 spikes.
Life Sci Alliance, 6, 2023
|
|
8H0X
 
 | | Structure of SARS-CoV-1 Spike Protein with Engineered x1 Disulfide (S370C and D967C), Locked-1 Conformation | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, BILIVERDINE IX ALPHA, LINOLEIC ACID, ... | | 著者 | Zhang, X, Li, Z, Liu, Y, Wang, J, Fu, L, Wang, P, He, J, Xiong, X. | | 登録日 | 2022-09-30 | | 公開日 | 2022-11-09 | | 最終更新日 | 2023-07-19 | | 実験手法 | ELECTRON MICROSCOPY (2.57 Å) | | 主引用文献 | Disulfide stabilization reveals conserved dynamic features between SARS-CoV-1 and SARS-CoV-2 spikes.
Life Sci Alliance, 6, 2023
|
|
