4G6G
 
 | | Crystal structure of NDH with TRT | | 分子名称: | FLAVIN-ADENINE DINUCLEOTIDE, FRAGMENT OF TRITON X-100, MAGNESIUM ION, ... | | 著者 | Li, W, Feng, Y, Ge, J, Yang, M. | | 登録日 | 2012-07-19 | | 公開日 | 2012-10-24 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.39 Å) | | 主引用文献 | Structural insight into the type-II mitochondrial NADH dehydrogenases.
Nature, 491, 2012
|
|
4G74
 
 | | Crystal structure of NDH with Quinone | | 分子名称: | 2,3-DIMETHOXY-5-METHYL-6-(3,11,15,19-TETRAMETHYL-EICOSA-2,6,10,14,18-PENTAENYL)-[1,4]BENZOQUINONE, FLAVIN-ADENINE DINUCLEOTIDE, FRAGMENT OF TRITON X-100, ... | | 著者 | Li, W, Feng, Y, Ge, J, Yang, M. | | 登録日 | 2012-07-19 | | 公開日 | 2012-10-24 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.48 Å) | | 主引用文献 | Structural insight into the type-II mitochondrial NADH dehydrogenases.
Nature, 491, 2012
|
|
4G73
 
 | | Crystal structure of NDH with NADH and Quinone | | 分子名称: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 2,3-DIMETHOXY-5-METHYL-6-(3,11,15,19-TETRAMETHYL-EICOSA-2,6,10,14,18-PENTAENYL)-[1,4]BENZOQUINONE, FLAVIN-ADENINE DINUCLEOTIDE, ... | | 著者 | Li, W, Feng, Y, Ge, J, Yang, M. | | 登録日 | 2012-07-19 | | 公開日 | 2012-10-24 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.522 Å) | | 主引用文献 | Structural insight into the type-II mitochondrial NADH dehydrogenases.
Nature, 491, 2012
|
|
4PL3
 
 | | Crystal structure of murine IRE1 in complex with MKC9989 inhibitor | | 分子名称: | 7-hydroxy-6-methoxy-3-[2-(2-methoxyethoxy)ethyl]-4,8-dimethyl-2H-chromen-2-one, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | 著者 | Sanches, M, Duffy, N, Talukdar, M, Thevakumaran, N, Chiovitti, D, Al-awar, R, Patterson, J.B, Sicheri, F. | | 登録日 | 2014-05-16 | | 公開日 | 2014-09-03 | | 最終更新日 | 2024-11-13 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Structure and mechanism of action of the hydroxy-aryl-aldehyde class of IRE1 endoribonuclease inhibitors.
Nat Commun, 5, 2014
|
|
4PL4
 
 | | Crystal structure of murine IRE1 in complex with OICR464 inhibitor | | 分子名称: | 2,5,8,11,14,17,20,23,26,29,32,35,38,41,44,47,50,53,56,59,62,65,68,71,74,77,80-HEPTACOSAOXADOOCTACONTAN-82-OL, 2-methoxy-6-methyl-4-(4-methyl-3,4-dihydro-2H-1,4-benzoxazin-7-yl)phenol, ADENOSINE-5'-DIPHOSPHATE, ... | | 著者 | Sanches, M, Duffy, N, Talukdar, M, Thevakumaran, N, Chiovitti, D, Al-awar, R, Patterson, J.B, Sicheri, F. | | 登録日 | 2014-05-16 | | 公開日 | 2014-09-03 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Structure and mechanism of action of the hydroxy-aryl-aldehyde class of IRE1 endoribonuclease inhibitors.
Nat Commun, 5, 2014
|
|
4PL5
 
 | | Crystal structure of murine IRE1 in complex with OICR573 inhibitor | | 分子名称: | 2,5,8,11,14,17,20,23,26,29,32,35,38,41,44,47,50,53,56,59,62,65,68,71,74,77,80-HEPTACOSAOXADOOCTACONTAN-82-OL, 3-methoxy-5-methyl-4'-(morpholin-4-yl)biphenyl-4-ol, ADENOSINE-5'-DIPHOSPHATE, ... | | 著者 | Sanches, M, Duffy, N, Talukdar, M, Thevakumaran, N, Chiovitti, D, Al-awar, R, Patterson, J.B, Sicheri, F. | | 登録日 | 2014-05-16 | | 公開日 | 2014-09-03 | | 最終更新日 | 2024-11-13 | | 実験手法 | X-RAY DIFFRACTION (3.4 Å) | | 主引用文献 | Structure and mechanism of action of the hydroxy-aryl-aldehyde class of IRE1 endoribonuclease inhibitors.
Nat Commun, 5, 2014
|
|
3SKH
 
 | |
3SKA
 
 | |
3TYV
 
 | |
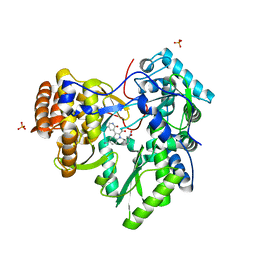
3TYQ
 
 | |
8GT6
 
 | | human STING With agonist HB3089 | | 分子名称: | 1-[(2E)-4-{5-carbamoyl-2-[(1-ethyl-3-methyl-1H-pyrazole-5-carbonyl)amino]-7-[3-(morpholin-4-yl)propoxy]-1H-benzimidazol-1-yl}but-2-en-1-yl]-2-[(1-ethyl-3-methyl-1H-pyrazole-5-carbonyl)amino]-7-methyl-1H-furo[3,2-e]benzimidazole-5-carboxamide, Stimulator of interferon genes protein | | 著者 | Wang, Z, Yu, X. | | 登録日 | 2022-09-07 | | 公開日 | 2022-12-28 | | 最終更新日 | 2024-06-19 | | 実験手法 | ELECTRON MICROSCOPY (3.47 Å) | | 主引用文献 | Structural insights into a shared mechanism of human STING activation by a potent agonist and an autoimmune disease-associated mutation.
Cell Discov, 8, 2022
|
|
8GSZ
 
 | | Structure of STING SAVI-related mutant V147L | | 分子名称: | Stimulator of interferon genes protein | | 著者 | Wang, Z, Yu, X. | | 登録日 | 2022-09-07 | | 公開日 | 2022-12-28 | | 最終更新日 | 2024-06-19 | | 実験手法 | ELECTRON MICROSCOPY (3.65 Å) | | 主引用文献 | Structural insights into a shared mechanism of human STING activation by a potent agonist and an autoimmune disease-associated mutation.
Cell Discov, 8, 2022
|
|
8XXI
 
 | | Structure of y+LAT1 bound with Leu | | 分子名称: | LEUCINE, SODIUM ION, Y+L amino acid transporter 1 | | 著者 | Yan, R.H, Dai, L, Zhang, T. | | 登録日 | 2024-01-18 | | 公開日 | 2025-01-22 | | 最終更新日 | 2025-04-02 | | 実験手法 | ELECTRON MICROSCOPY (3.04 Å) | | 主引用文献 | Structural basis for the substrate recognition and transport mechanism of the human y + LAT1-4F2hc transporter complex.
Sci Adv, 11, 2025
|
|
8XYJ
 
 | | Structure of y+LAT1 bound with Lys | | 分子名称: | LYSINE, Y+L amino acid transporter 1 | | 著者 | Yan, R.H, Dai, L, Zhang, T. | | 登録日 | 2024-01-19 | | 公開日 | 2025-01-22 | | 最終更新日 | 2025-04-02 | | 実験手法 | ELECTRON MICROSCOPY (3.33 Å) | | 主引用文献 | Structural basis for the substrate recognition and transport mechanism of the human y + LAT1-4F2hc transporter complex.
Sci Adv, 11, 2025
|
|
8YLP
 
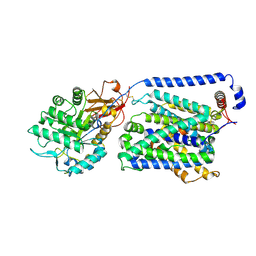 | | Overall structure of the y+LAT1-4F2hc bound with Arg | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ARGININE, Amino acid transporter heavy chain SLC3A2, ... | | 著者 | Yan, R.H, Dai, L, Zhang, Y.Y, Zhang, T. | | 登録日 | 2024-03-06 | | 公開日 | 2025-03-12 | | 最終更新日 | 2025-04-02 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | Structural basis for the substrate recognition and transport mechanism of the human y + LAT1-4F2hc transporter complex.
Sci Adv, 11, 2025
|
|
5JIL
 
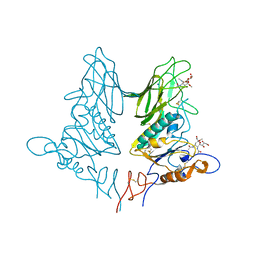 | | Crystal structure of rat coronavirus strain New-Jersey Hemagglutinin-Esterase in complex with 4N-acetyl sialic acid | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin-esterase, ... | | 著者 | Bakkers, M.J.G, Feitsma, L.J, de Groot, R.J, Huizinga, E.G. | | 登録日 | 2016-04-22 | | 公開日 | 2016-05-11 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Coronavirus receptor switch explained from the stereochemistry of protein-carbohydrate interactions and a single mutation.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
8I4L
 
 | | Capsid structure of the Cyanophage P-SCSP1u | | 分子名称: | The capsid protein(gp 19) of P-SCSP1u | | 著者 | Liu, H, Dang, S. | | 登録日 | 2023-01-19 | | 公開日 | 2023-10-25 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (3.23 Å) | | 主引用文献 | Cryo-EM structure of cyanophage P-SCSP1u offers insights into DNA gating and evolution of T7-like viruses.
Nat Commun, 14, 2023
|
|
8I4M
 
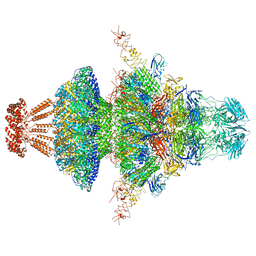 | | Portal-tail complex structure of the Cyanophage P-SCSP1u | | 分子名称: | Adaptor protein(gp22) of the cyanophage P-SCSP1u, Fiber protein(gp 28) of the cyanophage P-SCSP1u, Nozzle protein(gp 23) of the cyanophage P-SCSP1u, ... | | 著者 | Liu, H, Dang, S. | | 登録日 | 2023-01-19 | | 公開日 | 2023-11-01 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (3.81 Å) | | 主引用文献 | Cryo-EM structure of cyanophage P-SCSP1u offers insights into DNA gating and evolution of T7-like viruses.
Nat Commun, 14, 2023
|
|
7XG7
 
 | | Crystal structure of PstS protein from cyanophage Syn19 | | 分子名称: | GLYCEROL, PHOSPHATE ION, Phosphate transporter subunit | | 著者 | Cai, K, Jiang, Y.L. | | 登録日 | 2022-04-03 | | 公開日 | 2023-02-08 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Biochemical and structural characterization of the cyanophage-encoded phosphate-binding protein: implications for enhanced phosphate uptake of infected cyanobacteria
Environ.Microbiol., 24, 2022
|
|
