2AOG
 
 | | Crystal structure analysis of HIV-1 protease mutant V82A with a substrate analog P2-NC | | 分子名称: | ACETIC ACID, GLYCEROL, HIV-1 PROTEASE (RETROPEPSIN), ... | | 著者 | Tie, Y, Boross, P.I, Wang, Y.F, Gaddis, L, Liu, F, Chen, X, Tozser, J, Harrison, R.W, Weber, I.T. | | 登録日 | 2005-08-12 | | 公開日 | 2006-01-17 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (1.1 Å) | | 主引用文献 | Molecular basis for substrate recognition and drug resistance from 1.1 to 1.6 angstroms resolution crystal structures of HIV-1 protease mutants with substrate analogs.
Febs J., 272, 2005
|
|
2AVS
 
 | | kinetics, stability, and structural changes in high resolution crystal structures of HIV-1 protease with drug resistant mutations L24I, I50V, and G73S | | 分子名称: | ACETIC ACID, DIMETHYL SULFOXIDE, N-[2(R)-HYDROXY-1(S)-INDANYL]-5-[(2(S)-TERTIARY BUTYLAMINOCARBONYL)-4(3-PYRIDYLMETHYL)PIPERAZINO]-4(S)-HYDROXY-2(R)-PHENYLMETHYLPENTANAMIDE, ... | | 著者 | Liu, F, Boross, P.I, Wang, Y.F, Tozser, J, Louis, J.M, Harrison, R.W, Weber, I.T. | | 登録日 | 2005-08-30 | | 公開日 | 2006-01-24 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (1.1 Å) | | 主引用文献 | Kinetic, stability, and structural changes in high-resolution crystal structures of HIV-1 protease with drug-resistant mutations L24I, I50V, and G73S.
J.Mol.Biol., 354, 2005
|
|
2AOH
 
 | | Crystal structure analysis of HIV-1 Protease mutant V82A with a substrate analog P6-PR | | 分子名称: | CHLORIDE ION, PEPTIDE INHIBITOR, POL POLYPROTEIN, ... | | 著者 | Tie, Y, Boross, P.I, Wang, Y.F, Gaddis, L, Liu, F, Chen, X, Tozser, J, Harrison, R.W, Weber, I.T. | | 登録日 | 2005-08-12 | | 公開日 | 2006-01-17 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (1.42 Å) | | 主引用文献 | Molecular basis for substrate recognition and drug resistance from 1.1 to 1.6 angstroms resolution crystal structures of HIV-1 protease mutants with substrate analogs.
Febs J., 272, 2005
|
|
7C7P
 
 | | Crystal structure of the SARS-CoV-2 main protease in complex with Telaprevir | | 分子名称: | (1S,3aR,6aS)-2-[(2S)-2-({(2S)-2-cyclohexyl-2-[(pyrazin-2-ylcarbonyl)amino]acetyl}amino)-3,3-dimethylbutanoyl]-N-[(2R,3S)-1-(cyclopropylamino)-2-hydroxy-1-oxohexan-3-yl]octahydrocyclopenta[c]pyrrole-1-carboxamide, 3C-like proteinase, CHLORIDE ION | | 著者 | Zeng, R, Qiao, J.X, Wang, Y.F, Li, Y.S, Yao, R, Yang, S.Y, Lei, J. | | 登録日 | 2020-05-26 | | 公開日 | 2020-07-29 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (1.74 Å) | | 主引用文献 | SARS-CoV-2 M pro inhibitors with antiviral activity in a transgenic mouse model.
Science, 371, 2021
|
|
7COM
 
 | | Crystal structure of the SARS-CoV-2 main protease in complex with Boceprevir (space group P212121) | | 分子名称: | 3C-like proteinase, boceprevir (bound form) | | 著者 | Zeng, R, Qiao, J.X, Wang, Y.F, Li, Y.S, Yao, R, Liu, J.M, Zhou, Y.L, Chen, P, Yang, S.Y, Lei, J. | | 登録日 | 2020-08-04 | | 公開日 | 2020-08-19 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | SARS-CoV-2 M pro inhibitors with antiviral activity in a transgenic mouse model.
Science, 371, 2021
|
|
3VF5
 
 | | Crystal Structure of HIV-1 Protease Mutant I47V with novel P1'-Ligands GRL-02031 | | 分子名称: | (3aS,5R,6aR)-hexahydro-2H-cyclopenta[b]furan-5-yl [(1S,2R)-1-benzyl-2-hydroxy-3-([(4-methoxyphenyl)sulfonyl]{[(2R)-5-oxopyrrolidin-2-yl]methyl}amino)propyl]carbamate, ACETATE ION, CHLORIDE ION, ... | | 著者 | Yu, X.X, Wang, Y.F, Chang, Y.C.E, Weber, I.T. | | 登録日 | 2012-01-09 | | 公開日 | 2012-11-21 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.25 Å) | | 主引用文献 | Potent antiviral HIV-1 protease inhibitor GRL-02031 adapts to the structures of drug resistant mutants with its P1'-pyrrolidinone ring.
J.Med.Chem., 55, 2012
|
|
3VF7
 
 | | Crystal Structure of HIV-1 Protease Mutant L76V with novel P1'-Ligands GRL-02031 | | 分子名称: | (3aS,5R,6aR)-hexahydro-2H-cyclopenta[b]furan-5-yl [(1S,2R)-1-benzyl-2-hydroxy-3-([(4-methoxyphenyl)sulfonyl]{[(2R)-5-oxopyrrolidin-2-yl]methyl}amino)propyl]carbamate, CHLORIDE ION, GLYCEROL, ... | | 著者 | Yu, X.X, Wang, Y.F, Chang, Y.C.E, Weber, I.T. | | 登録日 | 2012-01-09 | | 公開日 | 2012-11-21 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | Potent antiviral HIV-1 protease inhibitor GRL-02031 adapts to the structures of drug resistant mutants with its P1'-pyrrolidinone ring.
J.Med.Chem., 55, 2012
|
|
3VFA
 
 | | Crystal Structure of HIV-1 Protease Mutant V82A with novel P1'-Ligands GRL-02031 | | 分子名称: | (3aS,5R,6aR)-hexahydro-2H-cyclopenta[b]furan-5-yl [(1S,2R)-1-benzyl-2-hydroxy-3-([(4-methoxyphenyl)sulfonyl]{[(2R)-5-oxopyrrolidin-2-yl]methyl}amino)propyl]carbamate, CHLORIDE ION, SODIUM ION, ... | | 著者 | Yu, X.X, Wang, Y.F, Chang, Y.C.E, Weber, I.T. | | 登録日 | 2012-01-09 | | 公開日 | 2012-11-21 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.43 Å) | | 主引用文献 | Potent antiviral HIV-1 protease inhibitor GRL-02031 adapts to the structures of drug resistant mutants with its P1'-pyrrolidinone ring.
J.Med.Chem., 55, 2012
|
|
3VFB
 
 | | Crystal Structure of HIV-1 Protease Mutant N88D with novel P1'-Ligands GRL-02031 | | 分子名称: | (3aS,5R,6aR)-hexahydro-2H-cyclopenta[b]furan-5-yl [(1S,2R)-1-benzyl-2-hydroxy-3-([(4-methoxyphenyl)sulfonyl]{[(2R)-5-oxopyrrolidin-2-yl]methyl}amino)propyl]carbamate, CHLORIDE ION, GLYCEROL, ... | | 著者 | Yu, X.X, Wang, Y.F, Chang, Y.C.E, Weber, I.T. | | 登録日 | 2012-01-09 | | 公開日 | 2012-11-21 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Potent antiviral HIV-1 protease inhibitor GRL-02031 adapts to the structures of drug resistant mutants with its P1'-pyrrolidinone ring.
J.Med.Chem., 55, 2012
|
|
6DJ7
 
 | | HIV-1 protease with mutation L76V in complex with GRL-5010 (gem-difluoro-bis-tetrahydrofuran as P2 ligand) | | 分子名称: | (3R,3aS,6aS)-4,4-difluorohexahydrofuro[2,3-b]furan-3-yl [(2S,3R)-4-{[(4-aminophenyl)sulfonyl](2-methylpropyl)amino}-3-hydroxy-1-phenylbutan-2-yl]carbamate, ACETATE ION, CHLORIDE ION, ... | | 著者 | Wong-Sam, A.E, Wang, Y.F, Weber, I.T. | | 登録日 | 2018-05-24 | | 公開日 | 2018-10-17 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.31 Å) | | 主引用文献 | Drug Resistance Mutation L76V Alters Nonpolar Interactions at the Flap-Core Interface of HIV-1 Protease.
ACS Omega, 3, 2018
|
|
2IDW
 
 | | Crystal structure analysis of HIV-1 protease mutant V82A with a potent non-peptide inhibitor (UIC-94017) | | 分子名称: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, ACETIC ACID, CHLORIDE ION, ... | | 著者 | Tie, Y, Boross, P.I, Wang, Y.F, Gaddis, L, Manna, D, Hussain, A.K, Leshchenko, S, Ghosh, A.K, Louis, J.M, Harrison, R.W, Weber, I.T. | | 登録日 | 2006-09-15 | | 公開日 | 2006-10-03 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.1 Å) | | 主引用文献 | High Resolution Crystal Structures of HIV-1 Protease with a Potent Non-Peptide Inhibitor (Uic-94017) Active Against Multi-Drug-Resistant Clinical Strains.
J.Mol.Biol., 338, 2004
|
|
2IEO
 
 | | Crystal structure analysis of HIV-1 protease mutant I84V with a potent non-peptide inhibitor (UIC-94017) | | 分子名称: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, CHLORIDE ION, Protease, ... | | 著者 | Tie, Y, Boross, P.I, Wang, Y.F, Gaddis, L, Manna, D, Hussain, A.K, Leshchenko, S, Ghosh, A.K, Louis, J.M, Harrison, R.W, Weber, I.T. | | 登録日 | 2006-09-19 | | 公開日 | 2006-10-03 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.53 Å) | | 主引用文献 | High Resolution Crystal Structures of HIV-1 Protease with a Potent Non-Peptide Inhibitor (Uic-94017) Active Against Multi-Drug-Resistant Clinical Strains.
J.Mol.Biol., 338, 2004
|
|
6DJ5
 
 | | HIV-1 protease with mutation L76V in complex with GRL-0519 (tris-tetrahydrofuran as P2 ligand) | | 分子名称: | (3R,3aS,3bR,6aS,7aS)-octahydrodifuro[2,3-b:3',2'-d]furan-3-yl [(1S,2R)-1-benzyl-2-hydroxy-3-{[(4-methoxyphenyl)sulfonyl](2-methylpropyl)amino}propyl]carbamate, CHLORIDE ION, GLYCEROL, ... | | 著者 | Wong-Sam, A.E, Wang, Y.F, Weber, I.T. | | 登録日 | 2018-05-24 | | 公開日 | 2018-10-17 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Drug Resistance Mutation L76V Alters Nonpolar Interactions at the Flap-Core Interface of HIV-1 Protease.
ACS Omega, 3, 2018
|
|
2NMZ
 
 | | Crystal structure analysis of HIV-1 protease mutant V82A with a inhibitor saquinavir | | 分子名称: | (2S)-N-[(2S,3R)-4-[(2S,3S,4aS,8aS)-3-(tert-butylcarbamoyl)-3,4,4a,5,6,7,8,8a-octahydro-1H-isoquinolin-2-yl]-3-hydroxy-1 -phenyl-butan-2-yl]-2-(quinolin-2-ylcarbonylamino)butanediamide, PROTEASE, SULFATE ION | | 著者 | Tie, Y, Kovalevsky, A.Y, Boross, P, Wang, Y.F, Ghosh, A.K, Tozser, J, Harrison, R.W, Weber, I.T. | | 登録日 | 2006-10-23 | | 公開日 | 2007-03-13 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (0.97 Å) | | 主引用文献 | Atomic resolution crystal structures of HIV-1 protease and mutants V82A and I84V with saquinavir.
Proteins, 67, 2007
|
|
2NNK
 
 | | Crystal structure analysis of HIV-1 protease mutant I84V with a inhibitor saquinavir | | 分子名称: | (2S)-N-[(2S,3R)-4-[(2S,3S,4aS,8aS)-3-(tert-butylcarbamoyl)-3,4,4a,5,6,7,8,8a-octahydro-1H-isoquinolin-2-yl]-3-hydroxy-1 -phenyl-butan-2-yl]-2-(quinolin-2-ylcarbonylamino)butanediamide, ACETIC ACID, CHLORIDE ION, ... | | 著者 | Tie, Y, Kovalevsky, A.Y, Boross, P, Wang, Y.F, Ghosh, A.K, Tozser, J, Harrison, R.W, Weber, I.T. | | 登録日 | 2006-10-24 | | 公開日 | 2007-03-13 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.25 Å) | | 主引用文献 | Atomic resolution crystal structures of HIV-1 protease and mutants V82A and I84V with saquinavir.
Proteins, 67, 2007
|
|
2NMY
 
 | | Crystal structure analysis of HIV-1 protease mutant V82A with a inhibitor saquinavir | | 分子名称: | (2S)-N-[(2S,3R)-4-[(2S,3S,4aS,8aS)-3-(tert-butylcarbamoyl)-3,4,4a,5,6,7,8,8a-octahydro-1H-isoquinolin-2-yl]-3-hydroxy-1 -phenyl-butan-2-yl]-2-(quinolin-2-ylcarbonylamino)butanediamide, CHLORIDE ION, PROTEASE, ... | | 著者 | Tie, Y, Kovalevsky, A.Y, Boross, P, Wang, Y.F, Ghosh, A.K, Tozser, J, Harrison, R.W, Weber, I.T. | | 登録日 | 2006-10-23 | | 公開日 | 2007-03-13 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.1 Å) | | 主引用文献 | Atomic resolution crystal structures of HIV-1 protease and mutants V82A and I84V with saquinavir.
Proteins, 67, 2007
|
|
2NNP
 
 | | Crystal structure analysis of HIV-1 protease mutant I84V with a inhibitor saquinavir | | 分子名称: | (2S)-N-[(2S,3R)-4-[(2S,3S,4aS,8aS)-3-(tert-butylcarbamoyl)-3,4,4a,5,6,7,8,8a-octahydro-1H-isoquinolin-2-yl]-3-hydroxy-1 -phenyl-butan-2-yl]-2-(quinolin-2-ylcarbonylamino)butanediamide, ACETIC ACID, GLYCEROL, ... | | 著者 | Tie, Y, Kovalevsky, A.Y, Boross, P, Wang, Y.F, Ghosh, A.K, Tozser, J, Harrison, R.W, Weber, I.T. | | 登録日 | 2006-10-24 | | 公開日 | 2007-03-13 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.2 Å) | | 主引用文献 | Atomic resolution crystal structures of HIV-1 protease and mutants V82A and I84V with saquinavir.
Proteins, 67, 2007
|
|
7C8B
 
 | | Crystal structure of the SARS-CoV-2 main protease in complex with Z-VAD(OMe)-FMK | | 分子名称: | 3C-like proteinase, CHLORIDE ION, Z-VAD(OMe)-FMK | | 著者 | Zeng, R, Qiao, J.X, Wang, Y.F, Li, Y.S, Yao, R, Yang, S.Y, Lei, J. | | 登録日 | 2020-05-29 | | 公開日 | 2020-09-02 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Crystal structure of the SARS-CoV-2 main protease in complex with Z-VAD(OMe)-FMK
To Be Published
|
|
7CX9
 
 | | Crystal structure of the SARS-CoV-2 main protease in complex with INZ-1 | | 分子名称: | 3-iodanyl-1~{H}-indazole-7-carbaldehyde, 3C-like proteinase, CHLORIDE ION, ... | | 著者 | Zeng, R, Liu, X.L, Qiao, J.X, Nan, J.S, Wang, Y.F, Li, Y.S, Yang, S.Y, Lei, J. | | 登録日 | 2020-09-01 | | 公開日 | 2020-09-16 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.73 Å) | | 主引用文献 | Crystal structure of the SARS-CoV-2 main protease in complex with INZ-1
To Be Published
|
|
1AF6
 
 | | MALTOPORIN SUCROSE COMPLEX | | 分子名称: | MAGNESIUM ION, MALTOPORIN, beta-D-fructofuranose-(2-1)-alpha-D-glucopyranose | | 著者 | Dutzler, R, Schirmer, T. | | 登録日 | 1997-03-21 | | 公開日 | 1998-03-25 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Channel specificity: structural basis for sugar discrimination and differential flux rates in maltoporin.
J.Mol.Biol., 272, 1997
|
|
1MPQ
 
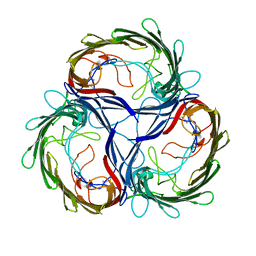 | | MALTOPORIN TREHALOSE COMPLEX | | 分子名称: | MALTOPORIN, alpha-D-glucopyranose-(1-1)-alpha-D-glucopyranose | | 著者 | Dutzler, R, Schirmer, T. | | 登録日 | 1997-03-24 | | 公開日 | 1998-03-25 | | 最終更新日 | 2024-10-09 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Channel specificity: structural basis for sugar discrimination and differential flux rates in maltoporin.
J.Mol.Biol., 272, 1997
|
|
7WDF
 
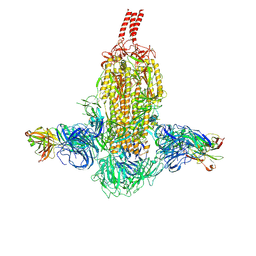 | |
7WD8
 
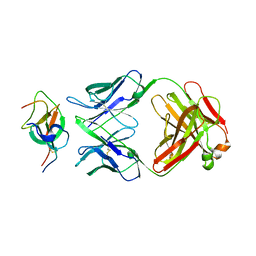 | |
7WCZ
 
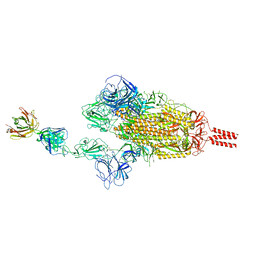 | |
7WD0
 
 | |
