1OGY
 
 | | Crystal structure of the heterodimeric nitrate reductase from Rhodobacter sphaeroides | | 分子名称: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, DIHEME CYTOCHROME C NAPB MOLECULE: NITRATE REDUCTASE, HEME C, ... | | 著者 | Arnoux, P, Sabaty, M, Alric, J, Frangioni, B, Guigliarelli, B, Adriano, J.-M, Pignol, D. | | 登録日 | 2003-05-19 | | 公開日 | 2003-10-09 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | Structural and Redox Plasticity in the Heterodimeric Periplasmic Nitrate Reductase
Nat.Struct.Biol., 10, 2003
|
|
8XLM
 
 | | Structure of the SARS-CoV-2 EG.5.1 spike glycoprotein in complex with ACE2 (1-up state) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | 著者 | Nomai, T, Anraku, Y, Kita, S, Hashiguchi, T, Maenaka, K. | | 登録日 | 2023-12-26 | | 公開日 | 2024-05-01 | | 最終更新日 | 2024-11-13 | | 実験手法 | ELECTRON MICROSCOPY (3.22 Å) | | 主引用文献 | Virological characteristics of the SARS-CoV-2 Omicron EG.5.1 variant.
Microbiol Immunol, 68, 2024
|
|
8XLN
 
 | | Structure of the SARS-CoV-2 EG.5.1 spike RBD in complex with ACE2 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | 著者 | Nomai, T, Anraku, Y, Kita, S, Hashiguchi, T, Maenaka, K. | | 登録日 | 2023-12-26 | | 公開日 | 2024-05-01 | | 最終更新日 | 2024-10-23 | | 実験手法 | ELECTRON MICROSCOPY (3.78 Å) | | 主引用文献 | Virological characteristics of the SARS-CoV-2 Omicron EG.5.1 variant.
Microbiol Immunol, 68, 2024
|
|
8WMD
 
 | | Structure of the SARS-CoV-2 EG.5.1 spike glycoprotein (closed-2 state) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | 著者 | Nomai, T, Anraku, Y, Kita, S, Hashiguchi, T, Maenaka, K. | | 登録日 | 2023-10-03 | | 公開日 | 2024-04-24 | | 最終更新日 | 2024-11-06 | | 実験手法 | ELECTRON MICROSCOPY (2.71 Å) | | 主引用文献 | Virological characteristics of the SARS-CoV-2 Omicron EG.5.1 variant.
Microbiol Immunol, 68, 2024
|
|
8WMF
 
 | | Structure of the SARS-CoV-2 EG.5.1 spike glycoprotein (closed-1 state) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | 著者 | Nomai, T, Anraku, Y, Kita, S, Hashiguchi, T, Maenaka, K. | | 登録日 | 2023-10-03 | | 公開日 | 2024-04-24 | | 最終更新日 | 2024-10-16 | | 実験手法 | ELECTRON MICROSCOPY (2.51 Å) | | 主引用文献 | Virological characteristics of the SARS-CoV-2 Omicron EG.5.1 variant.
Microbiol Immunol, 68, 2024
|
|
7BNR
 
 | |
7BNK
 
 | |
6ENE
 
 | | OmpF orthologue from Enterobacter cloacae (OmpE35) | | 分子名称: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, Outer membrane protein (Porin), SULFATE ION, ... | | 著者 | van den Berg, B, Abellon-Ruiz, J, Basle, A. | | 登録日 | 2017-10-04 | | 公開日 | 2018-10-31 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Getting Drugs into Gram-Negative Bacteria: Rational Rules for Permeation through General Porins.
Acs Infect Dis., 4, 2018
|
|
6RYK
 
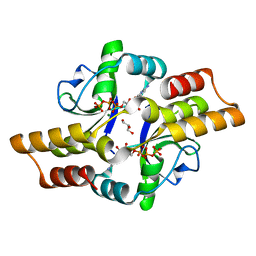 | |
7QAC
 
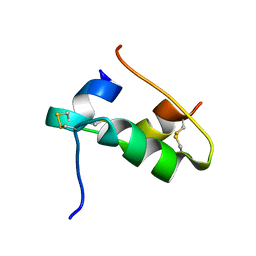 | | The T2 structure of polycrystalline cubic human insulin | | 分子名称: | Insulin A chain, Insulin B chain | | 著者 | Karavassili, F, Triandafillidis, D.P, Valmas, A, Spiliopoulou, M, Fili, S, Kontou, P, Bowler, M.W, Von Dreele, R.B, Fitch, A, Margiolaki, I. | | 登録日 | 2021-11-16 | | 公開日 | 2023-06-21 | | 最終更新日 | 2024-10-09 | | 実験手法 | POWDER DIFFRACTION (2.29 Å) | | 主引用文献 | The T 2 structure of polycrystalline cubic human insulin.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
4C67
 
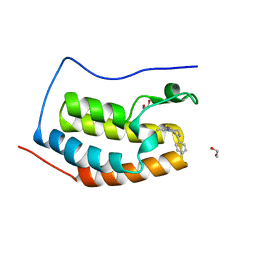 | | Discovery of Epigenetic Regulator I-BET762: Lead Optimization to Afford a Clinical Candidate Inhibitor of the BET Bromodomains | | 分子名称: | 1,2-ETHANEDIOL, 13-methyl-7-phenyl-3-thia-1,8,11,12-tetraazatricyclo trideca-2(6),4,7,10,12-pentaene, BROMODOMAIN-CONTAINING PROTEIN 4 | | 著者 | Chung, C, Mirguet, O. | | 登録日 | 2013-09-17 | | 公開日 | 2013-10-02 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Discovery of Epigenetic Regulator I-Bet762: Lead Optimization to Afford a Clinical Candidate Inhibitor of the Bet Bromodomains.
J.Med.Chem., 56, 2013
|
|
4C66
 
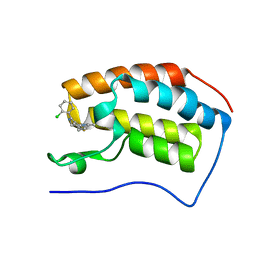 | | Discovery of Epigenetic Regulator I-BET762: Lead Optimization to Afford a Clinical Candidate Inhibitor of the BET Bromodomains | | 分子名称: | 4-(2-chlorophenyl)-2-ethyl-9-methyl-6,8-dihydrothieno[3,2-f][1,2,4]triazolo[4,3-a][1,4]diazepin-10-ium, BROMODOMAIN-CONTAINING PROTEIN 4 | | 著者 | Chung, C, Mirguet, O. | | 登録日 | 2013-09-17 | | 公開日 | 2013-10-02 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.87 Å) | | 主引用文献 | Discovery of Epigenetic Regulator I-Bet762: Lead Optimization to Afford a Clinical Candidate Inhibitor of the Bet Bromodomains.
J.Med.Chem., 56, 2013
|
|
4PUB
 
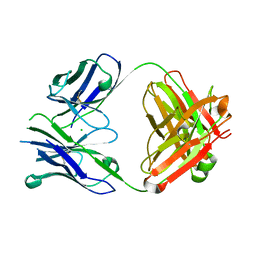 | | Crystal structure of Fab DX-2930 | | 分子名称: | CHLORIDE ION, DX-2930 HEAVY CHAIN, DX-2930 LIGHT CHAIN | | 著者 | Abendroth, J, Edwards, T.E, Nixon, A, Ladner, R. | | 登録日 | 2014-03-12 | | 公開日 | 2014-07-09 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Inhibition of plasma kallikrein by a highly specific active site blocking antibody.
J.Biol.Chem., 289, 2014
|
|
4OGX
 
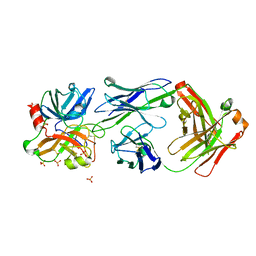 | | Crystal structure of Fab DX-2930 in complex with human plasma kallikrein at 2.4 Angstrom resolution | | 分子名称: | DX-2930 HEAVY CHAIN, DX-2930 LIGHT CHAIN, Plasma kallikrein, ... | | 著者 | Edwards, T.E, Clifton, M.C, Abendroth, J, Nixon, A, Ladner, R. | | 登録日 | 2014-01-16 | | 公開日 | 2014-07-09 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Inhibition of plasma kallikrein by a highly specific active site blocking antibody.
J.Biol.Chem., 289, 2014
|
|
7KPJ
 
 | |
2XJ4
 
 | |
2XJ9
 
 | |
