7L7E
 
 | |
7NZ9
 
 | |
7NZ7
 
 | |
7NZ8
 
 | |
7UBF
 
 | |
7UBD
 
 | |
7UBI
 
 | |
7UCP
 
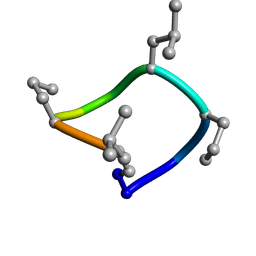 | | computationally designed macrocycle | | 分子名称: | computationally designed cyclic peptide D8.3.p2 | | 著者 | Bhardwaj, G, Baker, D, Rettie, S, Glynn, C, Sawaya, M. | | 登録日 | 2022-03-17 | | 公開日 | 2022-09-14 | | 最終更新日 | 2022-09-28 | | 実験手法 | X-RAY DIFFRACTION (0.85 Å) | | 主引用文献 | Accurate de novo design of membrane-traversing macrocycles.
Cell, 185, 2022
|
|
7UZL
 
 | |
7UBC
 
 | |
7UBE
 
 | |
7UBG
 
 | |
7UBH
 
 | |
7UWF
 
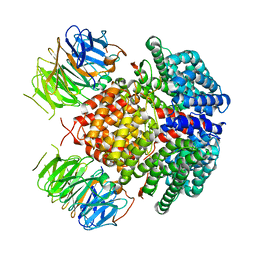 | | Human Rix1 sub-complex scaffold | | 分子名称: | Modulator of non-genomic activity of estrogen receptor, WD repeat-containing protein 18 | | 著者 | Gordon, J, Stanley, R.E. | | 登録日 | 2022-05-03 | | 公開日 | 2022-11-02 | | 最終更新日 | 2024-06-12 | | 実験手法 | ELECTRON MICROSCOPY (2.7 Å) | | 主引用文献 | Cryo-EM reveals the architecture of the PELP1-WDR18 molecular scaffold.
Nat Commun, 13, 2022
|
|
8CTO
 
 | |
8CUN
 
 | |
8CWA
 
 | |
1CY5
 
 | | CRYSTAL STRUCTURE OF THE APAF-1 CARD | | 分子名称: | BETA-MERCAPTOETHANOL, PROTEIN (APOPTOTIC PROTEASE ACTIVATING FACTOR 1), ZINC ION | | 著者 | Vaughn, D.E, Joshua-Tor, L. | | 登録日 | 1999-08-31 | | 公開日 | 1999-09-13 | | 最終更新日 | 2018-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | Crystal structure of Apaf-1 caspase recruitment domain: an alpha-helical Greek key fold for apoptotic signaling.
J.Mol.Biol., 293, 1999
|
|
6I9J
 
 | | Human transforming growth factor beta2 in a tetragonal crystal form | | 分子名称: | Transforming growth factor beta-2 proprotein | | 著者 | Gomis-Ruth, F.X, Marino-Puertas, L, del Amo-Maestro, L, Goulas, T. | | 登録日 | 2018-11-23 | | 公開日 | 2019-07-03 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Recombinant production, purification, crystallization, and structure analysis of human transforming growth factor beta 2 in a new conformation.
Sci Rep, 9, 2019
|
|
7QX4
 
 | | mosquitocidal Cry11Aa determined at pH 7 from naturally-occurring nanocrystals by Serial femtosecond crystallography | | 分子名称: | Pesticidal crystal protein Cry11Aa | | 著者 | De Zitter, E, Tetreau, G, Andreeva, E.A, Coquelle, N, Colletier, J.-P. | | 登録日 | 2022-01-26 | | 公開日 | 2022-07-27 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | De novo determination of mosquitocidal Cry11Aa and Cry11Ba structures from naturally-occurring nanocrystals.
Nat Commun, 13, 2022
|
|
7QX6
 
 | | mosquitocidal Cry11Aa-E583Q determined at pH 7 from naturally-occurring nanocrystals by Serial femtosecond crystallography | | 分子名称: | Pesticidal crystal protein Cry11Aa | | 著者 | De Zitter, E, Tetreau, G, Andreeva, E.A, Coquelle, N, Colletier, J.-P. | | 登録日 | 2022-01-26 | | 公開日 | 2022-07-27 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (3.3 Å) | | 主引用文献 | De novo determination of mosquitocidal Cry11Aa and Cry11Ba structures from naturally-occurring nanocrystals.
Nat Commun, 13, 2022
|
|
7QX5
 
 | | mosquitocidal Cry11Aa-Y449F determined at pH 7 from naturally-occurring nanocrystals by Serial femtosecond crystallography | | 分子名称: | Pesticidal crystal protein Cry11Aa | | 著者 | De Zitter, E, Tetreau, G, Andreeva, E.A, Coquelle, N, Colletier, J.-P. | | 登録日 | 2022-01-26 | | 公開日 | 2022-07-27 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | De novo determination of mosquitocidal Cry11Aa and Cry11Ba structures from naturally-occurring nanocrystals.
Nat Commun, 13, 2022
|
|
7QX7
 
 | | mosquitocidal Cry11Aa-F17Y determined at pH 7 from naturally-occurring nanocrystals by Serial femtosecond crystallography | | 分子名称: | Pesticidal crystal protein Cry11Aa | | 著者 | De Zitter, E, Tetreau, G, Andreeva, E.A, Coquelle, N, Colletier, J.-P. | | 登録日 | 2022-01-26 | | 公開日 | 2022-07-27 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (3.4 Å) | | 主引用文献 | De novo determination of mosquitocidal Cry11Aa and Cry11Ba structures from naturally-occurring nanocrystals.
Nat Commun, 13, 2022
|
|
5TY4
 
 | | MicroED structure of a complex between monomeric TGF-b and its receptor, TbRII, at 2.9 A resolution | | 分子名称: | TGF-beta receptor type-2, mmTGF-b2-7m | | 著者 | Weiss, S.C, de la Cruz, M.J, Hattne, J, Shi, D, Reyes, F.E, Callero, G, Gonen, T. | | 登録日 | 2016-11-18 | | 公開日 | 2017-04-26 | | 最終更新日 | 2023-10-04 | | 実験手法 | ELECTRON CRYSTALLOGRAPHY (2.9 Å) | | 主引用文献 | Atomic-resolution structures from fragmented protein crystals with the cryoEM method MicroED.
Nat. Methods, 14, 2017
|
|
8T84
 
 | | Racemic mixture of amyloid beta segment 35-MVGGVV-40 forms heterochiral rippled beta-sheet, includes hexafluoroisopropanol | | 分子名称: | 1,1,1,3,3,3-hexafluoropropan-2-ol, Racemic mixture of amyloid beta segment 35-MVGGVV-40 | | 著者 | Sawaya, M.R, Raskatov, J.A, Hazari, A. | | 登録日 | 2023-06-21 | | 公開日 | 2023-11-29 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.101 Å) | | 主引用文献 | Racemic Peptides from Amyloid beta and Amylin Form Rippled beta-Sheets Rather Than Pleated beta-Sheets.
J.Am.Chem.Soc., 145, 2023
|
|
