5UQP
 
 | | The crystal structure of cupin protein from Rhodococcus jostii RHA1 | | 分子名称: | CHLORIDE ION, Cupin, SULFATE ION, ... | | 著者 | Tan, K, Li, H, Clancy, S, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | 登録日 | 2017-02-08 | | 公開日 | 2017-02-22 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | The crystal structure of cupin protein from Rhodococcus jostii RHA1
To Be Published
|
|
9CCR
 
 | | Crystal structure of the EspE7 thioesterase mutant R35A from the esperamicin biosynthetic pathway at 1.6 A | | 分子名称: | DODECYL-COA, POTASSIUM ION, Thioesterase | | 著者 | Miller, M.D, Hankore, E.D, Xu, W, Kosgei, A.J, Bhardwaj, M, Thorson, J.S, Van Lanen, S.G, Phillips Jr, G.N. | | 登録日 | 2024-06-23 | | 公開日 | 2025-06-25 | | 実験手法 | X-RAY DIFFRACTION (1.57 Å) | | 主引用文献 | Functional and structural studies on the esperamicin thioesterase and progress toward understanding enediyne core biosynthesis
To Be Published
|
|
3DHG
 
 | | Crystal Structure of Toluene 4-Monoxygenase Hydroxylase | | 分子名称: | AZIDE ION, CALCIUM ION, FE (III) ION, ... | | 著者 | Bailey, L.J, Mccoy, J.G, Phillips Jr, G.N, Fox, B.G. | | 登録日 | 2008-06-17 | | 公開日 | 2008-12-30 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Structural consequences of effector protein complex formation in a diiron hydroxylase.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
3DCY
 
 | |
3DHH
 
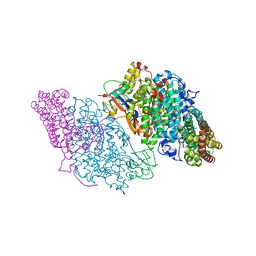 | | Crystal Structure of Resting State Toluene 4-Monoxygenase Hydroxylase Complexed with Effector Protein | | 分子名称: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4-BROMOPHENOL, CHLORIDE ION, ... | | 著者 | Bailey, L.J, Mccoy, J.G, Phillips Jr, G.N, Fox, B.G. | | 登録日 | 2008-06-17 | | 公開日 | 2008-12-30 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (1.94 Å) | | 主引用文献 | Structural consequences of effector protein complex formation in a diiron hydroxylase.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
3DHI
 
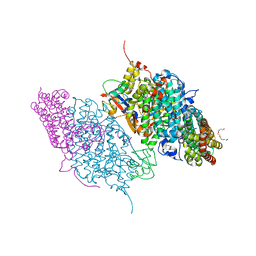 | | Crystal Structure of Reduced Toluene 4-Monoxygenase Hydroxylase Complexed with Effector Protein | | 分子名称: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, FE (III) ION, ... | | 著者 | Bailey, L.J, Mccoy, J.G, Phillips Jr, G.N, Fox, B.G. | | 登録日 | 2008-06-17 | | 公開日 | 2008-12-30 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (1.68 Å) | | 主引用文献 | Structural consequences of effector protein complex formation in a diiron hydroxylase.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
6P58
 
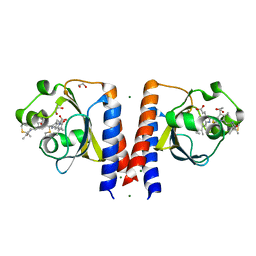 | | Dark and Steady State-Illuminated Crystal Structure of Cyanobacteriochrome Receptor PixJ at 150K | | 分子名称: | 1,2-ETHANEDIOL, MAGNESIUM ION, Methyl-accepting chemotaxis protein, ... | | 著者 | Clinger, J.A, Miller, M.D, Buirgie, E.S, Vierstra, R.D, Phillips Jr, G.N. | | 登録日 | 2019-05-29 | | 公開日 | 2019-12-18 | | 最終更新日 | 2024-10-09 | | 実験手法 | X-RAY DIFFRACTION (1.499 Å) | | 主引用文献 | Photoreversible interconversion of a phytochrome photosensory module in the crystalline state.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6P9V
 
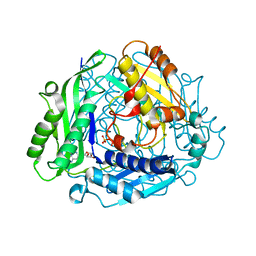 | | Crystal Structure of hMAT Mutant K289L | | 分子名称: | ADENOSINE, MAGNESIUM ION, POTASSIUM ION, ... | | 著者 | Miller, M.D, Xu, W, Huber, T.D, Clinger, J.A, Liu, Y, Thorson, J.S, Phillips Jr, G.N. | | 登録日 | 2019-06-10 | | 公開日 | 2020-04-22 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.051 Å) | | 主引用文献 | Methionine Adenosyltransferase Engineering to Enable Bioorthogonal Platforms for AdoMet-Utilizing Enzymes.
Acs Chem.Biol., 15, 2020
|
|
6PRY
 
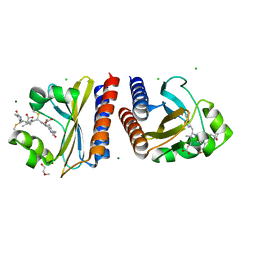 | | X-ray crystal structure of the blue-light absorbing state of PixJ from Thermosynechococcus elongatus by serial femtosecond crystallographic analysis | | 分子名称: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, ... | | 著者 | Burgie, E.S, Clinger, J.A, Miller, M.D, Phillips Jr, G.N, Vierstra, R.D, Orville, A.M, Kern, J.F. | | 登録日 | 2019-07-12 | | 公開日 | 2019-12-18 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Photoreversible interconversion of a phytochrome photosensory module in the crystalline state.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6PRU
 
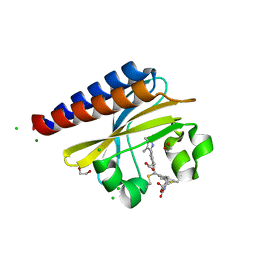 | | Photoconvertible crystals of PixJ from Thermosynechococcus elongatus | | 分子名称: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Burgie, E.S, Clinger, J.A, Miller, M.D, Phillips Jr, G.N, Vierstra, R.D. | | 登録日 | 2019-07-11 | | 公開日 | 2019-12-18 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (1.539 Å) | | 主引用文献 | Photoreversible interconversion of a phytochrome photosensory module in the crystalline state.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
4GLQ
 
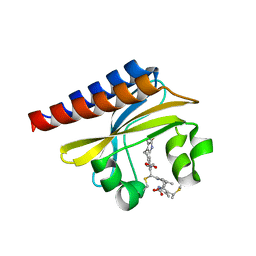 | | Crystal Structure of the blue-light absorbing form of the Thermosynechococcus elongatus PixJ GAF-domain | | 分子名称: | Methyl-accepting chemotaxis protein, Phycoviolobilin, blue light-absorbing form | | 著者 | Burgie, E.S, Walker, J.M, Phillips Jr, G.N, Vierstra, R.D. | | 登録日 | 2012-08-14 | | 公開日 | 2013-01-16 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (1.772 Å) | | 主引用文献 | A Photo-Labile Thioether Linkage to Phycoviolobilin Provides the Foundation for the Blue/Green Photocycles in DXCF-Cyanobacteriochromes.
Structure, 21, 2013
|
|
1ABS
 
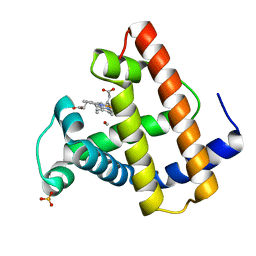 | | PHOTOLYSED CARBONMONOXY-MYOGLOBIN AT 20 K | | 分子名称: | CARBON MONOXIDE, MYOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE, ... | | 著者 | Schlichting, I, Berendzen, J, Phillips Jr, G.N, Sweet, R.M. | | 登録日 | 1997-01-28 | | 公開日 | 1997-04-01 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Crystal structure of photolysed carbonmonoxy-myoglobin.
Nature, 371, 1994
|
|
2H39
 
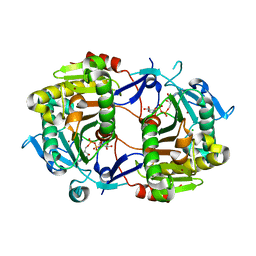 | | Crystal Structure of an ADP-Glucose Phosphorylase from Arabidopsis thaliana with bound ADP-Glucose | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE-GLUCOSE, CHLORIDE ION, Probable galactose-1-phosphate uridyl transferase, ... | | 著者 | McCoy, J.G, Wesenberg, G.E, Phillips Jr, G.N, Bitto, E, Bingman, C.A, Center for Eukaryotic Structural Genomics (CESG) | | 登録日 | 2006-05-22 | | 公開日 | 2006-06-13 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.23 Å) | | 主引用文献 | Crystal Structure of an ADP-Glucose Phosphorylase from Arabidopsis thaliana with bound ADP-Glucose
To be Published
|
|
6BBX
 
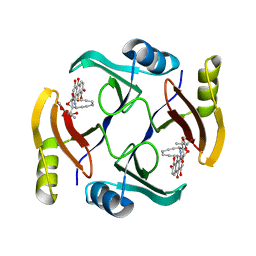 | | Crystal structure of TnmS3 in complex with TNM C | | 分子名称: | Glyoxalase/bleomycin resisance protein/dioxygenase, methyl (2R,3R)-2,3-dihydroxy-3-[(1aS,11S,11aR,14Z,18R)-3,7,8,18-tetrahydroxy-4,9-dioxo-4,9,10,11-tetrahydro-11aH-11,1a-hept[3]ene[1,5]diynonaphtho[2,3-h]oxireno[c]quinolin-11a-yl]butanoate | | 著者 | Chang, C.Y, Chang, C, Nocek, B, Rudolf, J.D, Joachimiak, A, Phillips Jr, G.N, Shen, B, Enzyme Discovery for Natural Product Biosynthesis (NatPro), Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2017-10-19 | | 公開日 | 2018-07-04 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Resistance to Enediyne Antitumor Antibiotics by Sequestration.
Cell Chem Biol, 25, 2018
|
|
4ZAS
 
 | | Crystal structure of sugar aminotransferase CalS13 from Micromonospora echinospora | | 分子名称: | CalS13, SULFATE ION, THYMIDINE-5'-DIPHOSPHATE, ... | | 著者 | Wang, F, Singh, S, Miller, M.D, Thorson, J.S, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | 登録日 | 2015-04-13 | | 公開日 | 2015-04-29 | | 最終更新日 | 2025-04-02 | | 実験手法 | X-RAY DIFFRACTION (2.47 Å) | | 主引用文献 | Structure characterization of sugar aminotransferases CalS13 and WecE provides the basis for a unifying structural model for stereochemical outcome.
To Be Published
|
|
4Z5P
 
 | | Crystal structure of the LnmA cytochrome P450 hydroxylase from the leinamycin biosynthetic pathway of Streptomyces atroolivaceus S-140 at 1.9 A resolution | | 分子名称: | Cytochrome P450 hydroxylase, PROTOPORPHYRIN IX CONTAINING FE, TRIETHYLENE GLYCOL | | 著者 | Ma, M, Lohman, J, Rudolf, J, Miller, M.D, Cao, H, Osipiuk, J, Babnigg, G, Phillips Jr, G.N, Joachimiak, A, Shen, B, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | 登録日 | 2015-04-02 | | 公開日 | 2015-07-29 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Crystal structure of the LnmA cytochrome P450 hydroxylase from the leinamycin biosynthetic pathway of Streptomyces atroolivaceus S-140
To be Published
|
|
4Z5Q
 
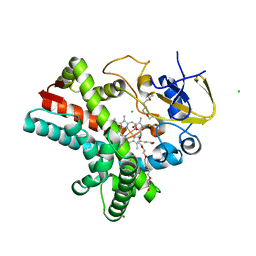 | | Crystal structure of the LnmZ cytochrome P450 hydroxylase from the leinamycin biosynthetic pathway of Streptomyces atroolivaceus S-140 at 1.8 A resolution | | 分子名称: | 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, CHLORIDE ION, Cytochrome P450 hydroxylase, ... | | 著者 | Ma, M, Lohman, J, Rudolf, J, Miller, M.D, Cao, H, Osipiuk, J, Joachimiak, A, Phillips Jr, G.N, Shen, B, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | 登録日 | 2015-04-02 | | 公開日 | 2015-08-12 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.801 Å) | | 主引用文献 | Crystal structure of the LnmZ cytochrome P450 hydroxylase from the leinamycin biosynthetic pathway of Streptomyces atroolivaceus S-140
To be Published
|
|
5CQF
 
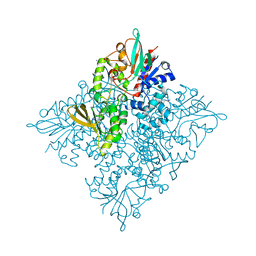 | | Crystal structure of L-lysine 6-monooxygenase from Pseudomonas syringae | | 分子名称: | IODIDE ION, L-lysine 6-monooxygenase | | 著者 | Michalska, K, Bigelow, L, Jedrzejczak, R, Weerth, R.S, Cao, H, Yennamalli, R, Phillips Jr, G.N, Thomas, M.G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | 登録日 | 2015-07-21 | | 公開日 | 2015-09-30 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (2.28 Å) | | 主引用文献 | Crystal structure of L-lysine 6-monooxygenase from Pseudomonas syringae
To Be Published
|
|
5UID
 
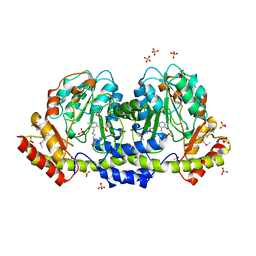 | | The crystal structure of an aminotransferase TlmJ from Streptoalloteichus hindustanus | | 分子名称: | Aminotransferase TlmJ, PYRIDOXAL-5'-PHOSPHATE, SULFATE ION | | 著者 | Tan, K, Bigelow, L, Bearden, J, Phillips Jr, G.N, Joachmiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | 登録日 | 2017-01-13 | | 公開日 | 2017-02-01 | | 最終更新日 | 2025-04-02 | | 実験手法 | X-RAY DIFFRACTION (2.18 Å) | | 主引用文献 | The crystal structure of an aminotransferase TlmJ from Streptoalloteichus hindustanus.
To Be Published
|
|
5UHJ
 
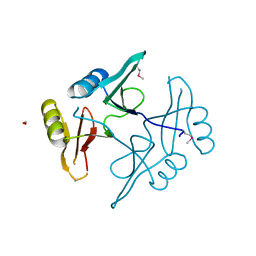 | | The crystal structure of a natural product biosynthetic enzyme from Streptomyces sp. CB03234 | | 分子名称: | FORMIC ACID, Glyoxalase/bleomycin resisance protein/dioxygenase | | 著者 | Tan, K, Li, H, Endres, M, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | 登録日 | 2017-01-11 | | 公開日 | 2017-01-25 | | 最終更新日 | 2024-11-20 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | The crystal structure of a natural product biosynthetic enzyme from Streptomyces sp. CB03234
To Be Published
|
|
5UMY
 
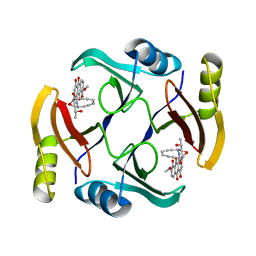 | | Crystal structure of TnmS3 in complex with tiancimycin | | 分子名称: | (1aS,11S,11aR,14Z,18R)-3,8,18-trihydroxy-11a-[(1R)-1-hydroxyethyl]-7-methoxy-11,11a-dihydro-4H-11,1a-hept[3]ene[1,5]diynonaphtho[2,3-h]oxireno[c]quinoline-4,9(10H)-dione, Glyoxalase/bleomycin resisance protein/dioxygenase | | 著者 | Chang, C.Y, Chang, C, Nocek, B, Rudolf, J.D, Joachimiak, A, Phillips Jr, G.N, SHen, B, Enzyme Discovery for Natural Product Biosynthesis (NatPro), Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2017-01-29 | | 公開日 | 2018-07-04 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.78 Å) | | 主引用文献 | Resistance to Enediyne Antitumor Antibiotics by Sequestration.
Cell Chem Biol, 25, 2018
|
|
5UJP
 
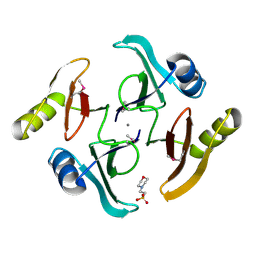 | | The crystal structure of a glyoxalase/bleomycin resistance protein from Streptomyces sp. CB03234 | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, Glyoxalase/bleomycin resisance protein/dioxygenase | | 著者 | Tan, K, Li, H, Endres, M, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | 登録日 | 2017-01-18 | | 公開日 | 2017-02-22 | | 最終更新日 | 2024-11-13 | | 実験手法 | X-RAY DIFFRACTION (1.42 Å) | | 主引用文献 | The crystal structure of a glyoxalase/bleomycin resistance protein from Streptomyces sp. CB03234
To Be Published
|
|
5UMP
 
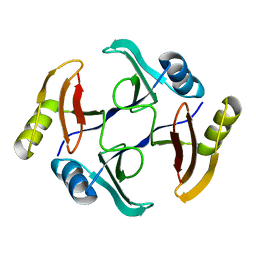 | | Crystal structure of TnmS3, an antibiotic binding protein from Streptomyces sp. CB03234 | | 分子名称: | Glyoxalase/bleomycin resisance protein/dioxygenase | | 著者 | Chang, C.Y, Chang, C, Nocek, B, Rudolf, J.D, Joachimiak, A, Phillips Jr, G.N, Shen, B, Enzyme Discovery for Natural Product Biosynthesis (NatPro), Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2017-01-29 | | 公開日 | 2018-07-04 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.08 Å) | | 主引用文献 | Resistance to Enediyne Antitumor Antibiotics by Sequestration.
Cell Chem Biol, 25, 2018
|
|
5UMW
 
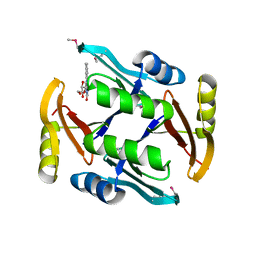 | | Crystal structure of TnmS2, an antibiotic binding protein from Streptomyces sp. CB03234 | | 分子名称: | Glyoxalase/bleomycin resisance protein/dioxygenase, RIBOFLAVIN | | 著者 | Chang, C.Y, Chang, C, Nocek, B, Rudolf, J.D, Joachimiak, A, Phillips Jr, G.N, Shen, B, Enzyme Discovery for Natural Product Biosynthesis (NatPro), Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2017-01-29 | | 公開日 | 2018-07-04 | | 最終更新日 | 2024-11-13 | | 実験手法 | X-RAY DIFFRACTION (2.27 Å) | | 主引用文献 | Resistance to Enediyne Antitumor Antibiotics by Sequestration.
Cell Chem Biol, 25, 2018
|
|
5VPJ
 
 | | The crystal structure of a thioesterase from Actinomadura verrucosospora | | 分子名称: | CHLORIDE ION, TETRAETHYLENE GLYCOL, Thioesterase | | 著者 | Tan, K, Joachimiak, G, Endres, M, Phillips Jr, G.N, Joachmiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | 登録日 | 2017-05-05 | | 公開日 | 2017-07-19 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | The crystal structure of a thioesterase from Actinomadura verrucosospora.
To Be Published
|
|
