7JVA
 
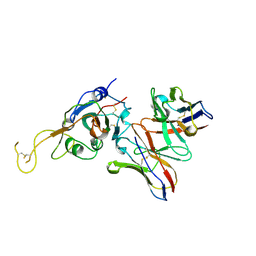 | | SARS-CoV-2 spike in complex with the S2A4 neutralizing antibody Fab fragment (local refinement of the receptor-binding domain and Fab variable domains) | | 分子名称: | S2A4 Fab heavy chain, S2A4 Fab light chain, Spike glycoprotein, ... | | 著者 | Park, Y.J, Tortorici, M.A, Walls, A.C, Czudnochowski, N, Seattle Structural Genomics Center for Infectious Disease (SSGCID), Snell, G, Veesler, D. | | 登録日 | 2020-08-20 | | 公開日 | 2020-10-14 | | 最終更新日 | 2021-01-27 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Mapping Neutralizing and Immunodominant Sites on the SARS-CoV-2 Spike Receptor-Binding Domain by Structure-Guided High-Resolution Serology.
Cell, 183, 2020
|
|
7K3Q
 
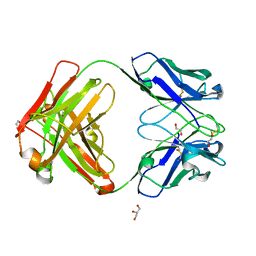 | | An ultra-potent human neutralizing antibody locks the SARS-CoV-2 spike in the closed conformation | | 分子名称: | 1,2-ETHANEDIOL, Fab fragment of S2E12 monoclonal antibody, heavy chain, ... | | 著者 | Snell, G, Czudnochowski, N, Ng, C. | | 登録日 | 2020-09-12 | | 公開日 | 2020-10-07 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.38 Å) | | 主引用文献 | Ultrapotent human antibodies protect against SARS-CoV-2 challenge via multiple mechanisms.
Science, 370, 2020
|
|
7JXD
 
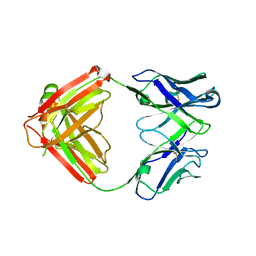 | | Mapping neutralizing and immunodominant sites on the SARS-CoV-2 spike receptor-binding domain by structure-guided high-resolution serology | | 分子名称: | S2A4 antigen-binding (Fab) fragment | | 著者 | Park, Y.J, Tortorici, M.A, Walls, A.C, Czudnochowski, N, Seattle Structural Genomics Center for Infectious Disease (SSGCID), Snell, G, Veesler, D. | | 登録日 | 2020-08-27 | | 公開日 | 2020-10-14 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Mapping Neutralizing and Immunodominant Sites on the SARS-CoV-2 Spike Receptor-Binding Domain by Structure-Guided High-Resolution Serology.
Cell, 183, 2020
|
|
7JXE
 
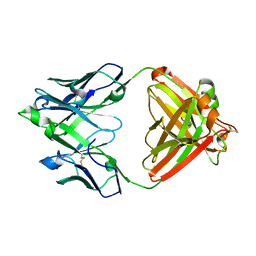 | | Mapping neutralizing and immunodominant sites on the SARS-CoV-2 spike receptor-binding domain by structure-guided high-resolution serology | | 分子名称: | S2X35 antigen-binding (Fab) fragment | | 著者 | Tortorici, M.A, Park, Y.J, Walls, A.C, Czudnochowski, N, Seattle Structural Genomics Center for Infectious Disease (SSGCID), Snell, G, Veesler, D. | | 登録日 | 2020-08-27 | | 公開日 | 2020-10-14 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.043 Å) | | 主引用文献 | Mapping Neutralizing and Immunodominant Sites on the SARS-CoV-2 Spike Receptor-Binding Domain by Structure-Guided High-Resolution Serology.
Cell, 183, 2020
|
|
7JVC
 
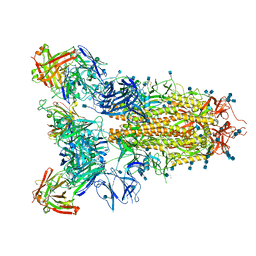 | | SARS-CoV-2 spike in complex with the S2A4 neutralizing antibody Fab fragment | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, S2A4 Fab heavy chain, ... | | 著者 | Park, Y.J, Tortorici, M.A, Walls, A.C, Czudnochowski, N, Seattle Structural Genomics Center for Infectious Disease (SSGCID), Snell, G, Veesler, D. | | 登録日 | 2020-08-20 | | 公開日 | 2020-10-14 | | 最終更新日 | 2021-06-23 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Mapping Neutralizing and Immunodominant Sites on the SARS-CoV-2 Spike Receptor-Binding Domain by Structure-Guided High-Resolution Serology.
Cell, 183, 2020
|
|
7JV6
 
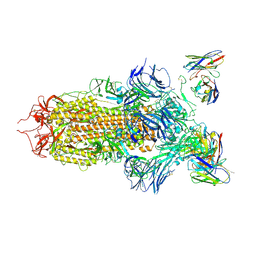 | | SARS-CoV-2 spike in complex with the S2H13 neutralizing antibody (closed conformation) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, S2H13 Fab heavy chain, ... | | 著者 | Park, Y.J, Tortorici, M.A, Walls, A.C, Czudnochowski, N, Seattle Structural Genomics Center for Infectious Disease (SSGCID), Snell, G, Veesler, D. | | 登録日 | 2020-08-20 | | 公開日 | 2020-10-14 | | 最終更新日 | 2021-06-23 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Mapping Neutralizing and Immunodominant Sites on the SARS-CoV-2 Spike Receptor-Binding Domain by Structure-Guided High-Resolution Serology.
Cell, 183, 2020
|
|
6LTV
 
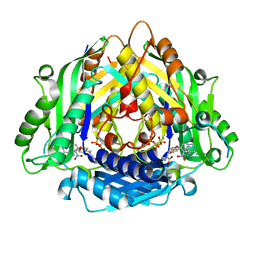 | | Crystal Structure of I122A/I330A variant of S-adenosylmethionine synthetase from Cryptosporidium hominis in complex with ONB-SAM (2-nitro benzyme S-adenosyl-methionine) | | 分子名称: | MAGNESIUM ION, S-adenosylmethionine synthase, TRIPHOSPHATE, ... | | 著者 | Singh, R.K, Michailidou, F, Rentmeister, A, Kuemmel, D. | | 登録日 | 2020-01-23 | | 公開日 | 2020-10-21 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.87 Å) | | 主引用文献 | Engineered SAM Synthetases for Enzymatic Generation of AdoMet Analogs with Photocaging Groups and Reversible DNA Modification in Cascade Reactions.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
6LTW
 
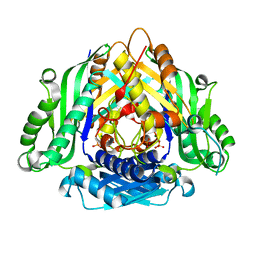 | | Crystal structure of Apo form of I122A/I330A variant of S-adenosylmethionine synthetase from Cryptosporidium hominis | | 分子名称: | MAGNESIUM ION, PHOSPHATE ION, S-adenosylmethionine synthase | | 著者 | Singh, R.K, Michailidou, F, Rentmeister, A, Kuemmel, D. | | 登録日 | 2020-01-23 | | 公開日 | 2020-10-21 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Engineered SAM Synthetases for Enzymatic Generation of AdoMet Analogs with Photocaging Groups and Reversible DNA Modification in Cascade Reactions.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
2REW
 
 | | Crystal Structure of PPARalpha ligand binding domain with BMS-631707 | | 分子名称: | (2S,3S)-1-(4-METHOXYPHENYL)-3-(3-(2-(5-METHYL-2-PHENYLOXAZOL-4-YL)ETHOXY)BENZYL)-4-OXOAZETIDINE-2-CARBOXYLIC ACID, N,N-BIS(3-D-GLUCONAMIDOPROPYL)DEOXYCHOLAMIDE, Peroxisome proliferator-activated receptor alpha | | 著者 | Muckelbauer, J. | | 登録日 | 2007-09-27 | | 公開日 | 2007-11-27 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | Discovery of Azetidinone Acids as Conformationally-Constrained Dual (alpha/gamma) PPAR Activators
To be Published
|
|
5VL7
 
 | | PCSK9 complex with Fab33 | | 分子名称: | Fab33 heavy chain, Fab33 light chain, Proprotein convertase subtilisin/kexin type 9 | | 著者 | Eigenbrot, C, Shia, S. | | 登録日 | 2017-04-25 | | 公開日 | 2017-08-16 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (3.5 Å) | | 主引用文献 | Discovery of a cryptic peptide-binding site on PCSK9 and design of antagonists.
Nat. Struct. Mol. Biol., 24, 2017
|
|
1HW5
 
 | | THE CAP/CRP VARIANT T127L/S128A | | 分子名称: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, CATABOLITE GENE ACTIVATOR | | 著者 | Chu, S.Y, Tordova, M, Gilliland, G.L, Gorshkova, I, Shi, Y. | | 登録日 | 2001-01-09 | | 公開日 | 2001-01-17 | | 最終更新日 | 2023-08-09 | | 実験手法 | X-RAY DIFFRACTION (1.82 Å) | | 主引用文献 | The structure of the T127L/S128A mutant of cAMP receptor protein facilitates promoter site binding
J.Biol.Chem., 276, 2001
|
|
3EZA
 
 | | COMPLEX OF THE AMINO TERMINAL DOMAIN OF ENZYME I AND THE HISTIDINE-CONTAINING PHOSPHOCARRIER PROTEIN HPR FROM ESCHERICHIA COLI NMR, RESTRAINED REGULARIZED MEAN STRUCTURE | | 分子名称: | HISTIDINE-CONTAINING PHOSPHOCARRIER PROTEIN HPR, PHOSPHOTRANSFERASE SYSTEM, ENZYME I | | 著者 | Clore, G.M, Garrett, D.S, Gronenborn, A.M. | | 登録日 | 1998-11-03 | | 公開日 | 1999-05-25 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of the 40,000 Mr phosphoryl transfer complex between the N-terminal domain of enzyme I and HPr.
Nat.Struct.Biol., 6, 1999
|
|
3EZB
 
 | | COMPLEX OF THE AMINO TERMINAL DOMAIN OF ENZYME I AND THE HISTIDINE-CONTAINING PHOSPHOCARRIER PROTEIN HPR FROM ESCHERICHIA COLI | | 分子名称: | PROTEIN (PHOSPHOCARRIER PROTEIN HPR), PROTEIN (PHOSPHOTRANSFER SYSTEM, ENZYME I) | | 著者 | Clore, G.M, Garrett, D.S, Gronenborn, A.M. | | 登録日 | 1998-11-03 | | 公開日 | 1999-12-16 | | 最終更新日 | 2023-12-27 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of the 40,000 Mr phosphoryl transfer complex between the N-terminal domain of enzyme I and HPr.
Nat.Struct.Biol., 6, 1999
|
|
3EZE
 
 | | COMPLEX OF THE AMINO TERMINAL DOMAIN OF ENZYME I AND THE HISTIDINE-CONTAINING PHOSPHOCARRIER PROTEIN HPR FROM ESCHERICHIA COLI NMR, RESTRAINED REGULARIZED MEAN STRUCTURE | | 分子名称: | PHOSPHITE ION, PROTEIN (PHOSPHOTRANSFERASE SYSTEM, ENZYME I), ... | | 著者 | Clore, G.M, Garrett, D.S, Gronenborn, A.M. | | 登録日 | 1998-11-04 | | 公開日 | 1998-12-16 | | 最終更新日 | 2023-12-27 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of the 40,000 Mr phosphoryl transfer complex between the N-terminal domain of enzyme I and HPr.
Nat.Struct.Biol., 6, 1999
|
|
4QT1
 
 | |
7R9W
 
 | | LC3A in complex with Fragment 1-1 | | 分子名称: | 4-phenoxybenzoic acid, GLYCEROL, Microtubule-associated proteins 1A/1B light chain 3A | | 著者 | Rouge, L, Steffek, M, Helgason, E, Dueber, E, Mulvihill, M. | | 登録日 | 2021-06-29 | | 公開日 | 2022-01-12 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | A Multifaceted Hit-Finding Approach Reveals Novel LC3 Family Ligands.
Biochemistry, 62, 2023
|
|
7R9Z
 
 | | LC3A in complex with Fragment 2-3 | | 分子名称: | (5-fluoro-1H-indol-3-yl)acetic acid, Microtubule-associated proteins 1A/1B light chain 3A | | 著者 | Rouge, L, Steffek, M, Helgason, E, Dueber, E, Mulvihill, M. | | 登録日 | 2021-06-29 | | 公開日 | 2022-01-12 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.72 Å) | | 主引用文献 | A Multifaceted Hit-Finding Approach Reveals Novel LC3 Family Ligands.
Biochemistry, 62, 2023
|
|
7RA0
 
 | | LC3A in complex with Fragment 2-10 | | 分子名称: | (5-ethyl-2-methyl-1H-indol-3-yl)acetic acid, Microtubule-associated proteins 1A/1B light chain 3A | | 著者 | Rouge, L, Steffek, M, Helgason, E, Dueber, E, Mulvihill, M. | | 登録日 | 2021-06-29 | | 公開日 | 2022-01-12 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.36 Å) | | 主引用文献 | A Multifaceted Hit-Finding Approach Reveals Novel LC3 Family Ligands.
Biochemistry, 62, 2023
|
|
1L3P
 
 | | CRYSTAL STRUCTURE OF THE FUNCTIONAL DOMAIN OF THE MAJOR GRASS POLLEN ALLERGEN Phl p 5b | | 分子名称: | MAGNESIUM ION, PHOSPHATE ION, POLLEN ALLERGEN Phl p 5b | | 著者 | Rajashankar, K.R, Bufe, A, Weber, W, Eschenburg, S, Lindner, B, Betzel, C. | | 登録日 | 2002-02-28 | | 公開日 | 2003-02-28 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.98 Å) | | 主引用文献 | Structure of the functional domain of the major grass-pollen allergen Phlp 5b.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
2HWG
 
 | | Structure of phosphorylated Enzyme I of the phosphoenolpyruvate:sugar phosphotransferase system | | 分子名称: | MAGNESIUM ION, OXALATE ION, Phosphoenolpyruvate-protein phosphotransferase | | 著者 | Lim, K, Teplyakov, A, Herzberg, O. | | 登録日 | 2006-08-01 | | 公開日 | 2006-11-14 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Structure of phosphorylated enzyme I, the phosphoenolpyruvate:sugar phosphotransferase system sugar translocation signal protein.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
6MV5
 
 | |
2EZA
 
 | | AMINO TERMINAL DOMAIN OF ENZYME I FROM ESCHERICHIA COLI, NMR, RESTRAINED REGULARIZED MEAN STRUCTURE | | 分子名称: | PHOSPHOTRANSFERASE SYSTEM, ENZYME I | | 著者 | Clore, G.M, Tjandra, N, Garrett, D.S, Gronenborn, A.M. | | 登録日 | 1997-05-07 | | 公開日 | 1997-08-20 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Defining long range order in NMR structure determination from the dependence of heteronuclear relaxation times on rotational diffusion anisotropy.
Nat.Struct.Biol., 4, 1997
|
|
2EZC
 
 | | AMINO TERMINAL DOMAIN OF ENZYME I FROM ESCHERICHIA COLI, NMR, 14 STRUCTURES | | 分子名称: | PHOSPHOTRANSFERASE SYSTEM, ENZYME I | | 著者 | Clore, G.M, Tjandra, N, Garrett, D.S, Gronenborn, A.M. | | 登録日 | 1997-05-07 | | 公開日 | 1997-08-20 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Defining long range order in NMR structure determination from the dependence of heteronuclear relaxation times on rotational diffusion anisotropy.
Nat.Struct.Biol., 4, 1997
|
|
2EZB
 
 | | AMINO TERMINAL DOMAIN OF ENZYME I FROM ESCHERICHIA COLI, NMR, 14 STRUCTURES | | 分子名称: | PHOSPHOTRANSFERASE SYSTEM, ENZYME I | | 著者 | Clore, G.M, Tjandra, N, Garrett, D.S, Gronenborn, A.M. | | 登録日 | 1997-05-07 | | 公開日 | 1997-08-20 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Defining long range order in NMR structure determination from the dependence of heteronuclear relaxation times on rotational diffusion anisotropy.
Nat.Struct.Biol., 4, 1997
|
|
1J6T
 
 | |
