5MCS
 
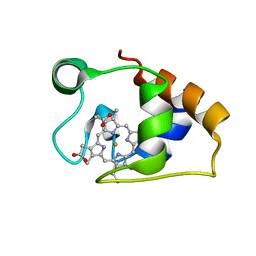 | | Solution structure and dynamics of the outer membrane cytochrome OmcF from Geobacter sulfurreducens | | 分子名称: | HEME C, Lipoprotein cytochrome c, 1 heme-binding site | | 著者 | Dantas, J.M, Silva, M.A, Morgado, L, Pantoja-Uceda, D, Turner, D.L, Bruix, M, Salgueiro, C.A. | | 登録日 | 2016-11-10 | | 公開日 | 2017-04-12 | | 最終更新日 | 2023-06-14 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure and dynamics of the outer membrane cytochrome OmcF from Geobacter sulfurreducens.
Biochim. Biophys. Acta, 1858, 2017
|
|
5MRG
 
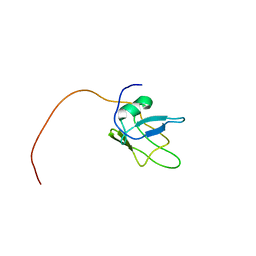 | | Solution structure of TDP-43 (residues 1-102) | | 分子名称: | TAR DNA-binding protein 43 | | 著者 | Mompean, M, Romano, V, Pantoja-Uceda, D, Stuani, C, Baralle, F.E, Laurents, D.V. | | 登録日 | 2016-12-22 | | 公開日 | 2017-06-07 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Point mutations in the N-terminal domain of transactive response DNA-binding protein 43 kDa (TDP-43) compromise its stability, dimerization, and functions.
J. Biol. Chem., 292, 2017
|
|
2LFK
 
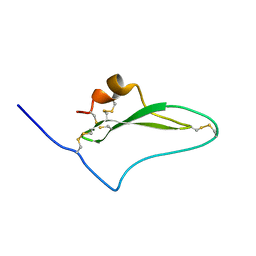 | | NMR solution structure of native TdPI-short | | 分子名称: | Tryptase inhibitor | | 著者 | Bronsoms, S, Pantoja-Uceda, D, Gabrijelcic-Geiger, D, Sanglas, L, Aviles, F, Santoro, J, Sommerhoff, C, Arolas, J. | | 登録日 | 2011-07-06 | | 公開日 | 2011-11-09 | | 最終更新日 | 2024-10-16 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Oxidative folding and structural analyses of a kunitz-related inhibitor and its disulfide intermediates: functional implications.
J.Mol.Biol., 414, 2011
|
|
2LFL
 
 | | NMR solution structure of the intermediate IIIb of TdPI-short | | 分子名称: | Tryptase inhibitor | | 著者 | Bronsoms, S, Pantoja-Uceda, D, Gabrijelcic-Geiger, D, Sanglas, L, Aviles, F, Santoro, J, Sommerhoff, C, Arolas, J. | | 登録日 | 2011-07-06 | | 公開日 | 2011-11-09 | | 最終更新日 | 2023-06-14 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Oxidative folding and structural analyses of a kunitz-related inhibitor and its disulfide intermediates: functional implications.
J.Mol.Biol., 414, 2011
|
|
2KG4
 
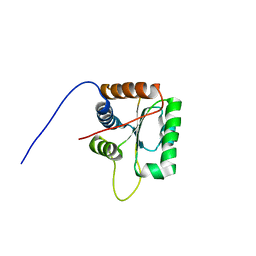 | | Three-dimensional structure of human Gadd45alpha in solution by NMR | | 分子名称: | Growth arrest and DNA-damage-inducible protein GADD45 alpha | | 著者 | Sanchez, R, Pantoja-Uceda, D, Prieto, J, Diercks, T, Campos-Olivas, R, Blanco, F.J. | | 登録日 | 2009-03-04 | | 公開日 | 2009-03-31 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of human growth arrest and DNA damage 45alpha (Gadd45alpha) and its interactions with proliferating cell nuclear antigen (PCNA) and Aurora A kinase
J.Biol.Chem., 285, 2010
|
|
4UHU
 
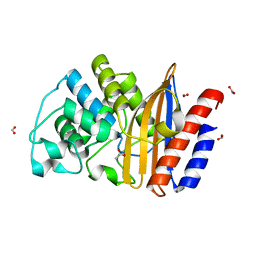 | | W229D mutant of the last common ancestor of Gram-negative bacteria (GNCA) beta-lactamase class A | | 分子名称: | ACETATE ION, FORMIC ACID, GNCA LACTAMASE W229D | | 著者 | Gavira, J.A, Risso, V.A, Martinez-Rodriguez, S, Sanchez-Ruiz, J.M. | | 登録日 | 2015-03-25 | | 公開日 | 2016-04-13 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.305 Å) | | 主引用文献 | De novo active sites for resurrected Precambrian enzymes.
Nat Commun, 8, 2017
|
|
2DCR
 
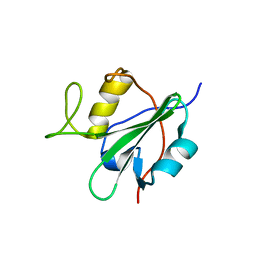 | |
2DCQ
 
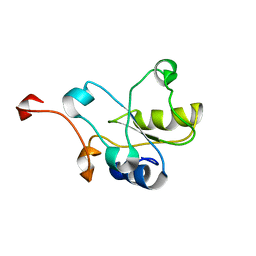 | |
4CNL
 
 | | Crystal structure of the Choline-binding domain of CbpL from Streptococcus pneumoniae | | 分子名称: | CHOLINE ION, GLYCEROL, PUTATIVE PNEUMOCOCCAL SURFACE PROTEIN, ... | | 著者 | Gutierrez-Fernandez, J, Bartual, S.G, Hermoso, J.A. | | 登録日 | 2014-01-23 | | 公開日 | 2015-02-18 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Modular Architecture and Unique Teichoic Acid Recognition Features of Choline-Binding Protein L (Cbpl) Contributing to Pneumococcal Pathogenesis.
Sci.Rep., 6, 2016
|
|
9FJN
 
 | | Solution NMR structure of a peptide encompassing residues 2-19 of the human formin INF2 | | 分子名称: | Inverted formin-2 | | 著者 | Jimenez, M.A, Comas, L, Labat-de-Hoz, L, Correas, I, Alonso, M.A. | | 登録日 | 2024-05-31 | | 公開日 | 2024-09-11 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structure and function of the N-terminal extension of the formin INF2.
Cell Mol Life Sci, 79, 2022
|
|
9FJW
 
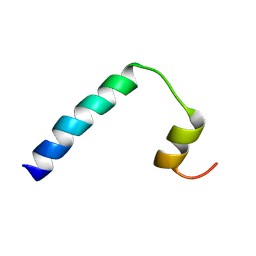 | | Solution NMR structure of a peptide encompassing residues 2-36 of the human formin INF2 | | 分子名称: | Inverted formin-2 | | 著者 | Jimenez, M.A, Comas, L, Labat-de-Hoz, L, Correas, I, Alonso, M.A. | | 登録日 | 2024-05-31 | | 公開日 | 2024-09-11 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structure and function of the N-terminal extension of the formin INF2.
Cell Mol Life Sci, 79, 2022
|
|
5FQQ
 
 | | Last common ancestor of Gram-negative bacteria (GNCA4) beta-lactamase class A | | 分子名称: | 2-(2-METHOXYETHOXY)ETHANOL, DI(HYDROXYETHYL)ETHER, GNCA4 LACTAMASE | | 著者 | Gavira, J.A, Martinez-Rodriguez, S, Risso, V.A, Sanchez-Ruiz, J.M. | | 登録日 | 2015-12-14 | | 公開日 | 2016-12-28 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.12 Å) | | 主引用文献 | De novo active sites for resurrected Precambrian enzymes.
Nat Commun, 8, 2017
|
|
5FQK
 
 | | W229D and F290W mutant of the last common ancestor of Gram-negative bacteria (GNCA4) beta-lactamase class A bound to 5(6)-nitrobenzotriazole (TS-analog) | | 分子名称: | 6-NITROBENZOTRIAZOLE, GNCA4 LACTAMASE W229D AND F290W | | 著者 | Gavira, J.A, Risso, V.A, Martinez-Rodriguez, S, Sanchez-Ruiz, J.M. | | 登録日 | 2015-12-11 | | 公開日 | 2016-12-21 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.767 Å) | | 主引用文献 | De novo active sites for resurrected Precambrian enzymes.
Nat Commun, 8, 2017
|
|
5FQM
 
 | | Last common ancestor of Gram Negative Bacteria (GNCA) Class A beta- lactamase | | 分子名称: | GLYCEROL, GNCA BETA LACTAMASE, SULFATE ION | | 著者 | Martinez Rodriguez, S, Gavira, J.A, Risso, V.A, Sanchez Ruiz, J.M. | | 登録日 | 2015-12-12 | | 公開日 | 2017-01-18 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | De novo active sites for resurrected Precambrian enzymes.
Nat Commun, 8, 2017
|
|
5FQI
 
 | | W229D and F290W mutant of the last common ancestor of Gram-negative bacteria (GNCA4) beta-lactamase class A | | 分子名称: | 1,2-ETHANEDIOL, 2-(2-METHOXYETHOXY)ETHANOL, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Gavira, J.A, Risso, V.A, Martinez-Rodriguez, S, Sanchez-Ruiz, J.M. | | 登録日 | 2015-12-11 | | 公開日 | 2016-12-21 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | De novo active sites for resurrected Precambrian enzymes.
Nat Commun, 8, 2017
|
|
5FQJ
 
 | | W229D mutant of the last common ancestor of Gram-negative bacteria (GNCA) beta-lactamase bound to 5(6)-nitrobenzotriazole (TS-analog) | | 分子名称: | 6-NITROBENZOTRIAZOLE, GNCA LACTAMASE W229D | | 著者 | Gavira, J.A, Martinez-Rodriguez, S, Risso, V.A, Sanchez-Ruiz, J.M. | | 登録日 | 2015-12-11 | | 公開日 | 2016-12-21 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.271 Å) | | 主引用文献 | De novo active sites for resurrected Precambrian enzymes.
Nat Commun, 8, 2017
|
|
2DCP
 
 | |
