7XWA
 
 | | Crystal structure of the receptor binding domain of SARS-CoV-2 Omicron BA.4/5 variant spike protein in complex with its receptor ACE2 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | 著者 | Suzuki, T, Kimura, K, Hashiguchi, T. | | 登録日 | 2022-05-26 | | 公開日 | 2022-09-28 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (3.36 Å) | | 主引用文献 | Virological characteristics of the SARS-CoV-2 Omicron BA.2 subvariants, including BA.4 and BA.5.
Cell, 185, 2022
|
|
1H8A
 
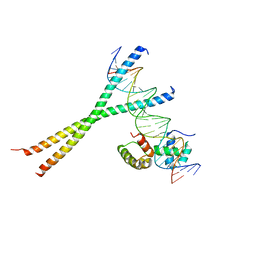 | | CRYSTAL STRUCTURE OF TERNARY PROTEIN-DNA COMPLEX3 | | 分子名称: | CAAT/ENHANCER BINDING PROTEIN BETA, DNA(5'-(*CP*CP*AP*GP*TP*CP*CP*GP*TP*TP*AP* AP*GP*GP*AP*TP*TP*GP*CP*GP*CP*CP*AP*CP*AP*T)-3'), DNA(5'-(*GP*AP*TP*GP*TP*GP*GP*CP*GP*CP*AP* AP*TP*CP*CP*TP*TP*AP*AP*CP*GP*GP*AP*CP*TP*G)-3'), ... | | 著者 | Tahirov, T.H, Ogata, K. | | 登録日 | 2001-01-31 | | 公開日 | 2002-01-28 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.23 Å) | | 主引用文献 | Mechanism of C-Myb-C/Ebpbeta Cooperation from Separated Sites on a Promoter
Cell(Cambridge,Mass.), 108, 2002
|
|
1H89
 
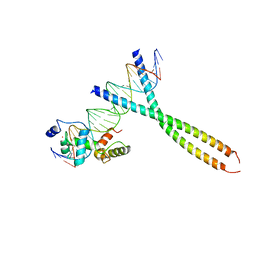 | | CRYSTAL STRUCTURE OF TERNARY PROTEIN-DNA COMPLEX2 | | 分子名称: | CAAT/ENHANCER BINDING PROTEIN BETA, DNA(5'-(*CP*CP*AP*GP*TP*CP*CP*GP*TP*TP*AP* AP*GP*GP*AP*TP*TP*GP*CP*GP*CP*CP*AP*CP*AP*T)-3'), DNA(5'-(*GP*AP*TP*GP*TP*GP*GP*CP*GP*CP*AP* AP*TP*CP*CP*TP*TP*AP*AP*CP*GP*GP*AP*CP*TP*G)-3'), ... | | 著者 | Tahirov, T.H, Ogata, K. | | 登録日 | 2001-01-30 | | 公開日 | 2002-01-28 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.45 Å) | | 主引用文献 | Mechanism of C-Myb-C/Ebpbeta Cooperation from Separated Sites on a Promoter
Cell(Cambridge,Mass.), 108, 2002
|
|
1H88
 
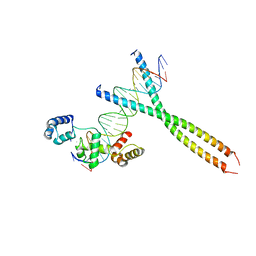 | | CRYSTAL STRUCTURE OF TERNARY PROTEIN-DNA COMPLEX1 | | 分子名称: | AMMONIUM ION, CCAAT/ENHANCER BINDING PROTEIN BETA, DNA(5'-(*CP*CP*AP*GP*TP*CP*CP*GP*TP*TP*AP* AP*GP*GP*AP*TP*TP*GP*CP*GP*CP*CP*AP*CP*AP*T)-3'), ... | | 著者 | Tahirov, T.H, Ogata, K. | | 登録日 | 2001-01-29 | | 公開日 | 2002-01-28 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Mechanism of C-Myb-C/Ebpbeta Cooperation from Separated Sites on a Promoter
Cell(Cambridge,Mass.), 108, 2002
|
|
1UCG
 
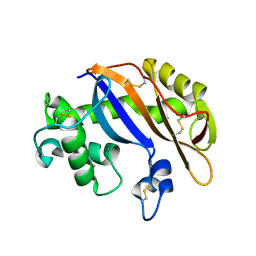 | | Crystal structure of Ribonuclease MC1 N71T mutant | | 分子名称: | MANGANESE (II) ION, Ribonuclease MC | | 著者 | Suzuki, A, Numata, T, Yao, M, Tanaka, I, Kimura, M. | | 登録日 | 2003-04-14 | | 公開日 | 2003-04-29 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Crystal structures of the ribonuclease MC1 mutants N71T and N71S in complex with 5'-GMP: structural basis for alterations in substrate specificity
Biochemistry, 42, 2003
|
|
1HJB
 
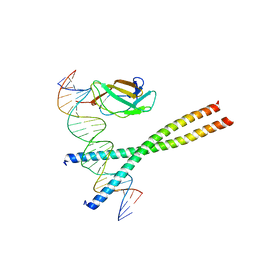 | |
1HJC
 
 | |
1IO4
 
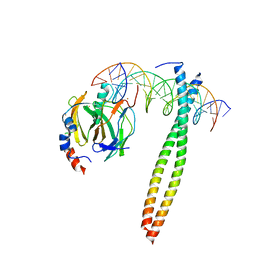 | |
6JZH
 
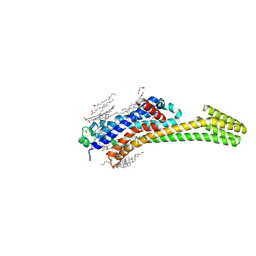 | | Structure of human A2A adenosine receptor in complex with ZM241385 obtained from SFX experiments under atmospheric pressure | | 分子名称: | 4-{2-[(7-amino-2-furan-2-yl[1,2,4]triazolo[1,5-a][1,3,5]triazin-5-yl)amino]ethyl}phenol, Adenosine receptor A2a,Soluble cytochrome b562,Adenosine receptor A2a, CHOLESTEROL, ... | | 著者 | Nango, E, Shimamura, T, Nakane, T, Yamanaka, Y, Mori, C, Kimura, K.T, Fujiwara, T, Tanaka, T, Iwata, S. | | 登録日 | 2019-05-02 | | 公開日 | 2019-10-30 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | High-viscosity sample-injection device for serial femtosecond crystallography at atmospheric pressure.
J.Appl.Crystallogr., 52, 2019
|
|
2ZYE
 
 | | Structure of HIV-1 Protease in Complex with Potent Inhibitor KNI-272 Determined by Neutron Crystallography | | 分子名称: | (4R)-N-tert-butyl-3-[(2S,3S)-2-hydroxy-3-({N-[(isoquinolin-5-yloxy)acetyl]-S-methyl-L-cysteinyl}amino)-4-phenylbutanoyl]-1,3-thiazolidine-4-carboxamide, protease | | 著者 | Adachi, M, Ohhara, T, Tamada, T, Okazaki, N, Kuroki, R. | | 登録日 | 2009-01-20 | | 公開日 | 2009-03-24 | | 最終更新日 | 2024-05-29 | | 実験手法 | NEUTRON DIFFRACTION (1.9 Å) | | 主引用文献 | Structure of HIV-1 protease in complex with potent inhibitor KNI-272 determined by high-resolution X-ray and neutron crystallography.
Proc.Natl.Acad.Sci.USA, 2009
|
|
7EXW
 
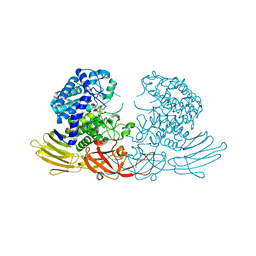 | | GH127 beta-L-arabinofuranosidase HypBA1 covalently complexed with alpha-L-arabinofuranosylamide | | 分子名称: | 2-bromanyl-N-[(2R,3R,4R,5S}-5-(hydroxymethyl)-3,4-bis(oxidanyl)oxolan-2-yl]ethanamide, Non-reducing end beta-L-arabinofuranosidase, ZINC ION | | 著者 | Sawano, K, Arakawa, T, Yamada, C, Fujita, K, Fushinobu, S. | | 登録日 | 2021-05-28 | | 公開日 | 2021-11-17 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Substrate complex structure, active site labeling and catalytic role of the zinc ion in cysteine glycosidase.
Glycobiology, 32, 2022
|
|
7EXU
 
 | | GH127 beta-L-arabinofuranosidase HypBA1 E322Q mutant complexed with p-nitrophenyl beta-L-arabinofuranoside | | 分子名称: | (2S,3R,4R,5R)-2-(hydroxymethyl)-5-(4-nitrophenoxy)oxolane-3,4-diol, Non-reducing end beta-L-arabinofuranosidase, ZINC ION | | 著者 | Maruyama, S, Arakawa, T, Yamada, C, Fujita, K, Fushinobu, S. | | 登録日 | 2021-05-28 | | 公開日 | 2021-11-17 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Substrate complex structure, active site labeling and catalytic role of the zinc ion in cysteine glycosidase.
Glycobiology, 32, 2022
|
|
7EXV
 
 | | GH127 beta-L-arabinofuranosidase HypBA1 covalently complexed with beta-L-arabinofuranoylamide | | 分子名称: | 2-bromanyl-N-[(2S,3R,4R,5S)-5-(hydroxymethyl)-3,4-bis(oxidanyl)oxolan-2-yl]ethanamide, Non-reducing end beta-L-arabinofuranosidase, ZINC ION | | 著者 | Sawano, K, Arakawa, T, Yamada, C, Fujita, K, Fushinobu, S. | | 登録日 | 2021-05-28 | | 公開日 | 2021-11-17 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Substrate complex structure, active site labeling and catalytic role of the zinc ion in cysteine glycosidase.
Glycobiology, 32, 2022
|
|
1WP6
 
 | | Crystal structure of maltohexaose-producing amylase from alkalophilic Bacillus sp.707. | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, Glucan 1,4-alpha-maltohexaosidase, ... | | 著者 | Kanai, R, Haga, K, Akiba, T, Yamane, K, Harata, K. | | 登録日 | 2004-08-31 | | 公開日 | 2004-11-30 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Biochemical and crystallographic analyses of maltohexaose-producing amylase from alkalophilic Bacillus sp. 707
Biochemistry, 43, 2004
|
|
1WPC
 
 | | Crystal structure of maltohexaose-producing amylase complexed with pseudo-maltononaose | | 分子名称: | 4,6-dideoxy-alpha-D-xylo-hexopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, 4,6-dideoxy-alpha-D-xylo-hexopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-beta-D-galactopyranose, 6-AMINO-4-HYDROXYMETHYL-CYCLOHEX-4-ENE-1,2,3-TRIOL, ... | | 著者 | Kanai, R, Haga, K, Akiba, T, Yamane, K, Harata, K. | | 登録日 | 2004-09-01 | | 公開日 | 2004-11-30 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Biochemical and crystallographic analyses of maltohexaose-producing amylase from alkalophilic Bacillus sp. 707
Biochemistry, 43, 2004
|
|
2ZAE
 
 | | Crystal structure of protein Ph1601p in complex with protein Ph1771p of archaeal ribonuclease P from Pyrococcus horikoshii OT3 | | 分子名称: | GLYCEROL, NITRATE ION, Ribonuclease P protein component 1, ... | | 著者 | Honda, T, Kakuta, Y, Kimura, M. | | 登録日 | 2007-10-04 | | 公開日 | 2008-10-14 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.21 Å) | | 主引用文献 | Structure of an archaeal homolog of the human protein complex Rpp21-Rpp29 that is a key core component for the assembly of active ribonuclease P.
J.Mol.Biol., 384, 2008
|
|
8JYM
 
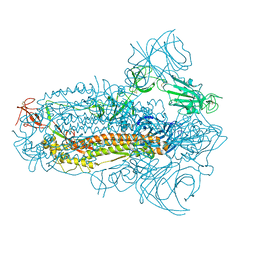 | | Structure of the SARS-CoV-2 XBB.1.5 spike glycoprotein (closed state 2) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | 著者 | Yajima, H, Anraku, Y, Kita, S, Kimura, K, Sasaki, J, Sasaki-Tabata, K, Maenaka, K, Hashiguchi, T. | | 登録日 | 2023-07-03 | | 公開日 | 2024-01-03 | | 実験手法 | ELECTRON MICROSCOPY (2.79 Å) | | 主引用文献 | Virological characteristics of the SARS-CoV-2 XBB.1.5 variant
To Be Published
|
|
8JYK
 
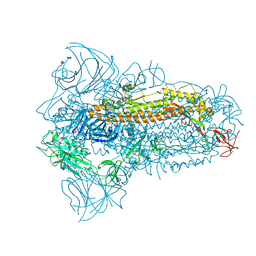 | | Structure of the SARS-CoV-2 XBB.1.5 spike glycoprotein (closed state 1) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | 著者 | Yajima, H, Anraku, Y, Kita, S, Kimura, K, Sasaki, J, Sasaki-Tabata, K, Maenaka, K, Hashiguchi, T. | | 登録日 | 2023-07-03 | | 公開日 | 2024-01-03 | | 実験手法 | ELECTRON MICROSCOPY (2.59 Å) | | 主引用文献 | Virological characteristics of the SARS-CoV-2 XBB.1.5 variant
To Be Published
|
|
8JYP
 
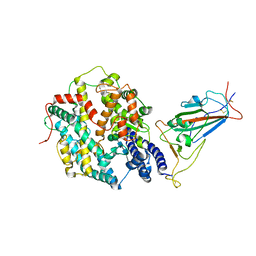 | | Structure of SARS-CoV-2 XBB.1.5 spike RBD in complex with ACE2 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | 著者 | Yajima, H, Anraku, Y, Kita, S, Kimura, K, Sasaki, J, Sasaki-Tabata, K, Maenaka, K, Hashiguchi, T. | | 登録日 | 2023-07-03 | | 公開日 | 2024-01-03 | | 実験手法 | ELECTRON MICROSCOPY (3.38 Å) | | 主引用文献 | Virological characteristics of the SARS-CoV-2 XBB.1.5 variant
To Be Published
|
|
8JYN
 
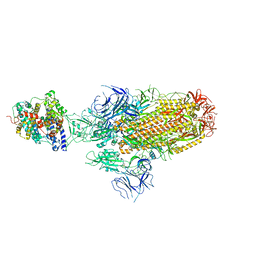 | | Structure of SARS-CoV-2 XBB.1.5 spike glycoprotein in complex with ACE2 (1-up state) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | 著者 | Yajima, H, Anraku, Y, Kita, S, Kimura, K, Sasaki, J, Sasaki-Tabata, K, Maenaka, K, Hashiguchi, T. | | 登録日 | 2023-07-03 | | 公開日 | 2024-01-03 | | 実験手法 | ELECTRON MICROSCOPY (3.04 Å) | | 主引用文献 | Virological characteristics of the SARS-CoV-2 XBB.1.5 variant
To Be Published
|
|
8JYO
 
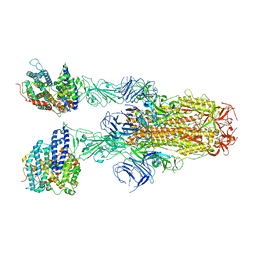 | | Structure of SARS-CoV-2 XBB.1.5 spike glycoprotein in complex with ACE2 (2-up state) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | 著者 | Yajima, H, Anraku, Y, Kita, S, Kimura, K, Sasaki, J, Sasaki-Tabata, K, Maenaka, K, Hashiguchi, T. | | 登録日 | 2023-07-03 | | 公開日 | 2024-01-03 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Virological characteristics of the SARS-CoV-2 XBB.1.5 variant
To Be Published
|
|
1GV5
 
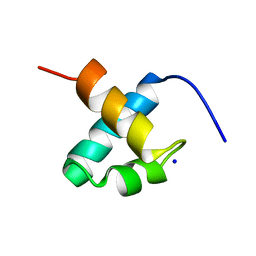 | | CRYSTAL STRUCTURE OF C-MYB R2 | | 分子名称: | MYB PROTO-ONCOGENE PROTEIN, SODIUM ION | | 著者 | Tahirov, T.H, Ogata, K. | | 登録日 | 2002-02-06 | | 公開日 | 2003-07-03 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.58 Å) | | 主引用文献 | Crystal Structure of C-Myb DNA-Binding Domain: Specific Na+ Binding and Correlation with NMR Structure
To be Published
|
|
1GVD
 
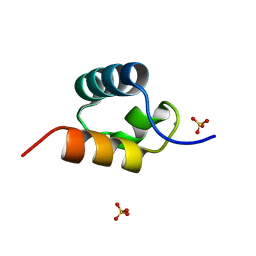 | | CRYSTAL STRUCTURE OF C-MYB R2 V103L MUTANT | | 分子名称: | AMMONIUM ION, MYB PROTO-ONCOGENE PROTEIN, SULFATE ION | | 著者 | Tahirov, T.H, Ogata, K. | | 登録日 | 2002-02-08 | | 公開日 | 2003-07-03 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | Crystal Structure of C-Myb DNA-Binding Domain: Specific Na+ Binding and Correlation with NMR Structure
To be Published
|
|
5B34
 
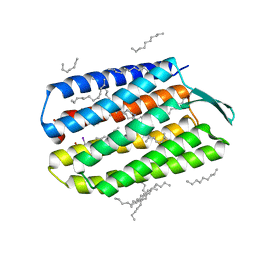 | | Serial Femtosecond Crystallography (SFX) of Ground State Bacteriorhodopsin Crystallized from Bicelles in Complex with Iodine-labeled Detergent HAD13a Determined Using 7-keV X-ray Free Electron Laser (XFEL) at SACLA | | 分子名称: | 2,4,6-tris(iodanyl)-5-(octanoylamino)benzene-1,3-dicarboxylic acid, Bacteriorhodopsin, DECANE, ... | | 著者 | Mizohata, E, Nakane, T. | | 登録日 | 2016-02-10 | | 公開日 | 2016-11-09 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Membrane protein structure determination by SAD, SIR, or SIRAS phasing in serial femtosecond crystallography using an iododetergent
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
7VDN
 
 | | High resolution crystal structure of Sperm Whale Myoglobin in the carbonmonoxy form | | 分子名称: | CARBON MONOXIDE, Myoglobin, PROTOPORPHYRIN IX CONTAINING FE, ... | | 著者 | Shibayama, N, Sato-Tomita, A, Ishimoto, N, Park, S.Y. | | 登録日 | 2021-09-07 | | 公開日 | 2022-09-14 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (0.93 Å) | | 主引用文献 | X-ray fluorescence holography of biological metal sites: Application to myoglobin.
Biochem.Biophys.Res.Commun., 635, 2022
|
|
