6XH2
 
 | |
6XLX
 
 | |
6XLV
 
 | |
6XKN
 
 | |
6XKO
 
 | |
4TU8
 
 | | STRUCTURE OF U2AF65 VARIANT WITH BRU5A6 DNA | | 分子名称: | 1,2-ETHANEDIOL, 1,4-DIETHYLENE DIOXIDE, DNA (5'-D(*UP*UP*UP*UP*(BRU)P*DA*U)-3'), ... | | 著者 | MCLAUGHLIN, K.J, JENKINS, J.L, Agrawal, A.A, KIELKOPF, C.L. | | 登録日 | 2014-06-24 | | 公開日 | 2014-11-26 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.918 Å) | | 主引用文献 | Structure-guided U2AF65 variant improves recognition and splicing of a defective pre-mRNA.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4TU7
 
 | | Structure of U2AF65 D231V variant with BrU5 DNA | | 分子名称: | 1,4-DIETHYLENE DIOXIDE, DNA (5'-D(*UP*UP*UP*UP*(BRU)P*UP*U)-3'), GLYCEROL, ... | | 著者 | Agrawal, A.A, Jenkins, J.L, Kielkopf, C.L. | | 登録日 | 2014-06-24 | | 公開日 | 2014-11-26 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.087 Å) | | 主引用文献 | Structure-guided U2AF65 variant improves recognition and splicing of a defective pre-mRNA.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
8FZA
 
 | |
1JN4
 
 | | The Crystal Structure of Ribonuclease A in complex with 2'-deoxyuridine 3'-pyrophosphate (P'-5') adenosine | | 分子名称: | ADENOSINE-5'-[TRIHYDROGEN DIPHOSPHATE] P'-3'-ESTER WITH 2'-DEOXYURIDINE, Pancreatic Ribonuclease A | | 著者 | Jardine, A.M, Leonidas, D.D, Jenkins, J.L, Park, C, Raines, R.T, Acharya, K.R, Shapiro, R. | | 登録日 | 2001-07-23 | | 公開日 | 2003-06-03 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Cleavage of 3',5'-Pyrophosphate-Linked Dinucleotides by Ribonuclease A and Angiogenin
Biochemistry, 40, 2001
|
|
8FB3
 
 | |
5EV1
 
 | | Structure I of Intact U2AF65 Recognizing a 3' Splice Site Signal | | 分子名称: | DI(HYDROXYETHYL)ETHER, DNA/RNA (5'-R(*UP*UP*U)-D(P*UP*UP*(BRU)P*U)-R(P*UP*U)-3'), SODIUM ION, ... | | 著者 | Agrawal, A.A, Jenkins, J.L, Kielkopf, C.L. | | 登録日 | 2015-11-19 | | 公開日 | 2016-02-24 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.037 Å) | | 主引用文献 | An extended U2AF(65)-RNA-binding domain recognizes the 3' splice site signal.
Nat Commun, 7, 2016
|
|
5EV2
 
 | | Structure II of Intact U2AF65 Recognizing the 3' Splice Site Signal | | 分子名称: | 1,4-DIETHYLENE DIOXIDE, DI(HYDROXYETHYL)ETHER, DNA (5'-R(P*UP*U)-D(P*UP*U)-R(P*U)-D(P*UP*(BRU)P*U)-3'), ... | | 著者 | Agrawal, A.A, Jenkins, J.L, Kielkopf, C.L. | | 登録日 | 2015-11-19 | | 公開日 | 2016-02-24 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.86 Å) | | 主引用文献 | An extended U2AF(65)-RNA-binding domain recognizes the 3' splice site signal.
Nat Commun, 7, 2016
|
|
5EV3
 
 | |
5EV4
 
 | |
4G6R
 
 | | Minimal Hairpin Ribozyme in the Transition State with G8I Variation | | 分子名称: | Loop A Ribozyme strand, Loop A Substrate strand, Loop B Ribozyme Strand, ... | | 著者 | Liberman, J.A, Jenkins, J.L, Krucinska, J, Wedekind, J.E. | | 登録日 | 2012-07-19 | | 公開日 | 2012-08-15 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.832 Å) | | 主引用文献 | A Transition-State Interaction Shifts Nucleobase Ionization toward Neutrality To Facilitate Small Ribozyme Catalysis.
J.Am.Chem.Soc., 134, 2012
|
|
4G6P
 
 | | Minimal Hairpin Ribozyme in the Precatalytic State with A38P Variation | | 分子名称: | COBALT HEXAMMINE(III), Loop A Substrate strand, Loop A and Loop B Ribozyme strand, ... | | 著者 | Liberman, J.A, Jenkins, J.L, Krucinska, J, Wedekind, J.E. | | 登録日 | 2012-07-19 | | 公開日 | 2012-08-15 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.641 Å) | | 主引用文献 | A Transition-State Interaction Shifts Nucleobase Ionization toward Neutrality To Facilitate Small Ribozyme Catalysis.
J.Am.Chem.Soc., 134, 2012
|
|
4G6S
 
 | | Minimal Hairpin Ribozyme in the Transition State with A38P Variation | | 分子名称: | COBALT HEXAMMINE(III), Loop A Substrate strand, Loop A and Loop B Ribozyme strand, ... | | 著者 | Liberman, J.A, Jenkins, J.L, Krucinska, J, Wedekind, J.E. | | 登録日 | 2012-07-19 | | 公開日 | 2012-08-08 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.84 Å) | | 主引用文献 | A Transition-State Interaction Shifts Nucleobase Ionization toward Neutrality To Facilitate Small Ribozyme Catalysis.
J.Am.Chem.Soc., 134, 2012
|
|
6XH1
 
 | |
6XLW
 
 | | Crystal structure of U2AF65 bound to AdML splice site sequence | | 分子名称: | DI(HYDROXYETHYL)ETHER, DNA/RNA (5'-R(P*UP*UP*(UD)P*UP*U)-D(P*(BRU))-R(P*CP*C)-3'), GLYCEROL, ... | | 著者 | Maji, D, Jenkins, J.L, Kielkopf, C.L. | | 登録日 | 2020-06-29 | | 公開日 | 2020-10-07 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Representative cancer-associated U2AF2 mutations alter RNA interactions and splicing.
J.Biol.Chem., 295, 2020
|
|
6N3D
 
 | | Structure of HIV Tat-specific factor 1 U2AF Homology Motif (APO-State) | | 分子名称: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | 著者 | Loerch, S, Jenkins, J.L, Kielkopf, C.L. | | 登録日 | 2018-11-15 | | 公開日 | 2019-01-02 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.13 Å) | | 主引用文献 | The pre-mRNA splicing and transcription factor Tat-SF1 is a functional partner of the spliceosome SF3b1 subunit via a U2AF homology motif interface.
J. Biol. Chem., 294, 2019
|
|
6N3F
 
 | | Structure of HIV Tat-specific factor 1 U2AF Homology Motif bound to SF3b1 ULM5 | | 分子名称: | DI(HYDROXYETHYL)ETHER, GLYCEROL, HIV Tat-specific factor 1, ... | | 著者 | Leach, J.R, Jenkins, J.L, Kielkopf, C.L. | | 登録日 | 2018-11-15 | | 公開日 | 2019-01-02 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.099 Å) | | 主引用文献 | The pre-mRNA splicing and transcription factor Tat-SF1 is a functional partner of the spliceosome SF3b1 subunit via a U2AF homology motif interface.
J. Biol. Chem., 294, 2019
|
|
6N3E
 
 | | Structure of HIV Tat-specific factor 1 U2AF Homology Motif bound to U2AF ligand motif 4 | | 分子名称: | FORMIC ACID, GLYCEROL, HIV Tat-specific factor 1, ... | | 著者 | Loerch, S, Jenkins, J.L, Kielkopf, C.L. | | 登録日 | 2018-11-15 | | 公開日 | 2019-01-02 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.893 Å) | | 主引用文献 | The pre-mRNA splicing and transcription factor Tat-SF1 is a functional partner of the spliceosome SF3b1 subunit via a U2AF homology motif interface.
J. Biol. Chem., 294, 2019
|
|
5SYT
 
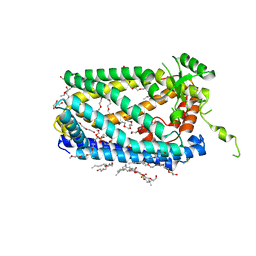 | | Crystal Structure of ZMPSTE24 | | 分子名称: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, CAAX prenyl protease 1 homolog, ... | | 著者 | Clark, K, Jenkins, J.L, Fedoriw, N, Dumont, M.E, Membrane Protein Structural Biology Consortium (MPSBC) | | 登録日 | 2016-08-11 | | 公開日 | 2016-11-02 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Human CaaX protease ZMPSTE24 expressed in yeast: Structure and inhibition by HIV protease inhibitors.
Protein Sci., 26, 2017
|
|
5W0H
 
 | |
2AY0
 
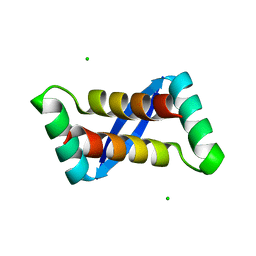 | | Structure of the Lys9Met mutant of the E. coli Proline Utilization A (PutA) DNA-binding domain. | | 分子名称: | Bifunctional putA protein, CHLORIDE ION | | 著者 | Larson, J.D, Schuermann, J.P, Zhou, Y, Jenkins, J.L, Becker, D.F, Tanner, J.J. | | 登録日 | 2005-09-06 | | 公開日 | 2006-08-15 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Crystal structures of the DNA-binding domain of Escherichia coli proline utilization A flavoprotein and analysis of the role of Lys9 in DNA recognition.
Protein Sci., 15, 2006
|
|
