7RXR
 
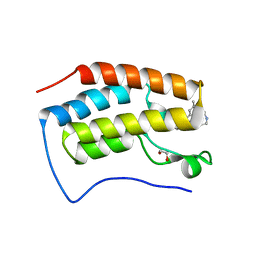 | | Crystal Structure of BRD4(D1) with 4-[4-(4-bromophenyl)-1-(piperidin-4-yl)-1H-imidazol-5-yl]-N-(3,5-dimethylphenyl)pyrimidin-2-amine | | 分子名称: | 1,2-ETHANEDIOL, 4-[4-(4-bromophenyl)-1-(piperidin-4-yl)-1H-imidazol-5-yl]-N-(3,5-dimethylphenyl)pyrimidin-2-amine, Bromodomain-containing protein 4 | | 著者 | Cui, H, Johnson, J.A, Shi, K, Aihara, H, Pomerantz, W.C.K. | | 登録日 | 2021-08-23 | | 公開日 | 2022-01-19 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.41 Å) | | 主引用文献 | A Structure-based Design Approach for Generating High Affinity BRD4 D1-Selective Chemical Probes.
J.Med.Chem., 65, 2022
|
|
7MC6
 
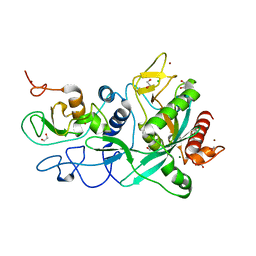 | | Crystal structure of the SARS-CoV-2 ExoN-nsp10 complex containing Mg2+ ion | | 分子名称: | 1,2-ETHANEDIOL, CHLORIDE ION, MAGNESIUM ION, ... | | 著者 | Moeller, N.M, Shi, K, Banerjee, S, Yin, L, Aihara, H. | | 登録日 | 2021-04-01 | | 公開日 | 2021-05-05 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structure and dynamics of SARS-CoV-2 proofreading exoribonuclease ExoN.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7MC5
 
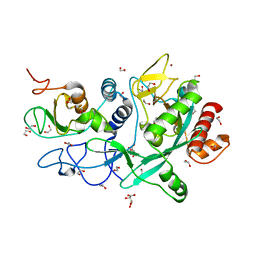 | | Crystal structure of the SARS-CoV-2 ExoN-nsp10 complex | | 分子名称: | 1,2-ETHANEDIOL, CHLORIDE ION, L(+)-TARTARIC ACID, ... | | 著者 | Moeller, N.M, Shi, K, Banerjee, S, Yin, L, Aihara, H. | | 登録日 | 2021-04-01 | | 公開日 | 2021-05-05 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.64 Å) | | 主引用文献 | Structure and dynamics of SARS-CoV-2 proofreading exoribonuclease ExoN.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7RWN
 
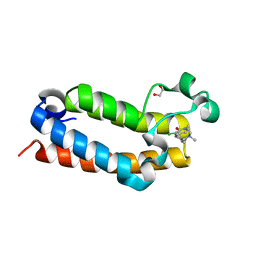 | | Crystal Structure of BPTF bromodomain in complex with 4-chloro-5-{4-[(dimethylamino)methyl]anilino}-2-methylpyridazin-3(2H)-one | | 分子名称: | 1,2-ETHANEDIOL, 4-chloro-5-{4-[(dimethylamino)methyl]anilino}-2-methylpyridazin-3(2H)-one, Nucleosome-remodeling factor subunit BPTF | | 著者 | Zahid, H, Buchholz, C, Johnson, J.A, Shi, K, Aihara, H, Pomerantz, W.C.K. | | 登録日 | 2021-08-20 | | 公開日 | 2022-08-24 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.39 Å) | | 主引用文献 | New Design Rules for Developing Potent Cell-Active Inhibitors of the Nucleosome Remodeling Factor (NURF) via BPTF Bromodomain Inhibition.
J.Med.Chem., 64, 2021
|
|
7RWP
 
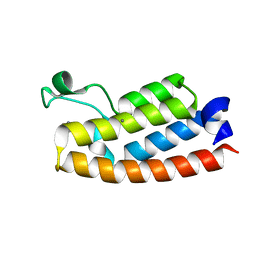 | | Crystal Structure of BPTF bromodomain in complex with 5-[4-(aminomethyl)anilino]-4-chloro-2-methylpyridazin-3(2H)-one | | 分子名称: | 5-[4-(aminomethyl)anilino]-4-chloro-2-methylpyridazin-3(2H)-one, CALCIUM ION, Nucleosome-remodeling factor subunit BPTF | | 著者 | Zahid, H, Buchholz, C, Johnson, J.A, Shi, K, Aihara, H, Pomerantz, W.C.K. | | 登録日 | 2021-08-20 | | 公開日 | 2022-08-24 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.73 Å) | | 主引用文献 | New Design Rules for Developing Potent Cell-Active Inhibitors of the Nucleosome Remodeling Factor (NURF) via BPTF Bromodomain Inhibition.
J.Med.Chem., 64, 2021
|
|
7RWQ
 
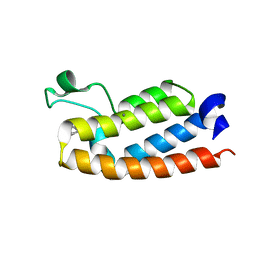 | | Crystal Structure of BPTF bromodomain in complex with 4-chloro-2-methyl-5-[(1,2,3,4-tetrahydroisoquinolin-6-yl)amino]pyridazin-3(2H)-one | | 分子名称: | 4-chloro-2-methyl-5-[(1,2,3,4-tetrahydroisoquinolin-6-yl)amino]pyridazin-3(2H)-one, CALCIUM ION, Nucleosome-remodeling factor subunit BPTF | | 著者 | Zahid, H, Buchholz, C, Johnson, J.A, Shi, K, Aihara, H, Pomerantz, W.C.K. | | 登録日 | 2021-08-20 | | 公開日 | 2022-08-24 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | New Design Rules for Developing Potent Cell-Active Inhibitors of the Nucleosome Remodeling Factor (NURF) via BPTF Bromodomain Inhibition.
J.Med.Chem., 64, 2021
|
|
7RWO
 
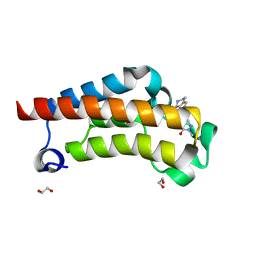 | | Crystal Structure of BPTF bromodomain in complex with 4-chloro-2-methyl-5-[(1,2,3,4-tetrahydroisoquinolin-7-yl)amino]pyridazin-3(2H)-one | | 分子名称: | 1,2-ETHANEDIOL, 4-chloro-2-methyl-5-[(1,2,3,4-tetrahydroisoquinolin-7-yl)amino]pyridazin-3(2H)-one, Nucleosome-remodeling factor subunit BPTF | | 著者 | Zahid, H, Buchholz, C, Johnson, J.A, Shi, K, Aihara, H, Pomerantz, W.C.K. | | 登録日 | 2021-08-20 | | 公開日 | 2022-08-24 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.58 Å) | | 主引用文献 | New Design Rules for Developing Potent Cell-Active Inhibitors of the Nucleosome Remodeling Factor (NURF) via BPTF Bromodomain Inhibition.
J.Med.Chem., 64, 2021
|
|
1C0N
 
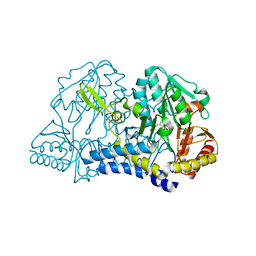 | | CSDB PROTEIN, NIFS HOMOLOGUE | | 分子名称: | ACETIC ACID, PROTEIN (CSDB PROTEIN), PYRIDOXAL-5'-PHOSPHATE | | 著者 | Fujii, T, Maeda, M, Mihara, H, Kurihara, T, Esaki, N, Hata, Y. | | 登録日 | 1999-07-17 | | 公開日 | 2000-07-17 | | 最終更新日 | 2017-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Structure of a NifS homologue: X-ray structure analysis of CsdB, an Escherichia coli counterpart of mammalian selenocysteine lyase
Biochemistry, 39, 2000
|
|
4E10
 
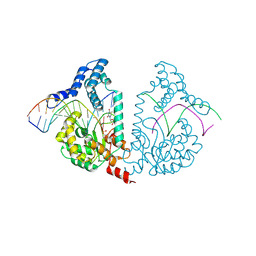 | | Protelomerase tela Y201A covalently complexed with substrate DNA | | 分子名称: | DNA (5'-D(*CP*AP*TP*AP*AP*(BRU)P*AP*AP*CP*AP*AP*(BRU)P*AP*T)-3'), DNA (5'-D(*CP*AP*TP*GP*AP*TP*AP*(BRU)P*(BRU)P*GP*(BRU)P*(BRU)P*AP*(BRU)P*(BRU)P*AP*(BRU)P*G)-3'), Protelomerase, ... | | 著者 | Shi, K, Aihara, H. | | 登録日 | 2012-03-05 | | 公開日 | 2013-02-13 | | 実験手法 | X-RAY DIFFRACTION (2.506 Å) | | 主引用文献 | An enzyme-catalyzed multistep DNA refolding mechanism in hairpin telomere formation.
Plos Biol., 11, 2013
|
|
1IQ5
 
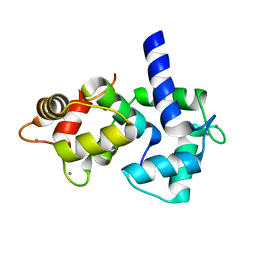 | | Calmodulin/nematode CA2+/Calmodulin dependent kinase kinase fragment | | 分子名称: | CA2+/CALMODULIN DEPENDENT KINASE KINASE, CALCIUM ION, CALMODULIN | | 著者 | Kurokawa, H, Osawa, M, Kurihara, H, Katayama, N, Tokumitsu, H, Swindells, M.B, Kainosho, M, Ikura, M. | | 登録日 | 2001-06-14 | | 公開日 | 2001-09-26 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Target-induced conformational adaptation of calmodulin revealed by the crystal structure of a complex with nematode Ca(2+)/calmodulin-dependent kinase kinase peptide
J.Mol.Biol., 312, 2001
|
|
5XFA
 
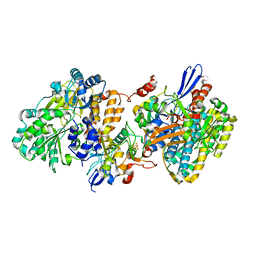 | | Crystal structure of NAD+-reducing [NiFe]-hydrogenase in the H2-reduced state | | 分子名称: | CARBONMONOXIDE-(DICYANO) IRON, FE2/S2 (INORGANIC) CLUSTER, IRON/SULFUR CLUSTER, ... | | 著者 | Shomura, Y, Taketa, M, Nakashima, H, Tai, H, Nakagawa, H, Ikeda, Y, Ishii, M, Igarashi, Y, Nishihara, H, Yoon, K.S, Ogo, S, Hirota, S, Higuchi, Y. | | 登録日 | 2017-04-09 | | 公開日 | 2017-08-23 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Structural basis of the redox switches in the NAD(+)-reducing soluble [NiFe]-hydrogenase
Science, 357, 2017
|
|
5XF9
 
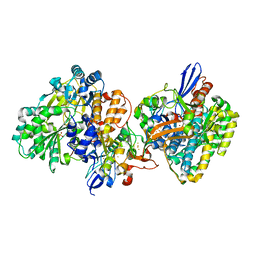 | | Crystal structure of NAD+-reducing [NiFe]-hydrogenase in the air-oxidized state | | 分子名称: | CARBONMONOXIDE-(DICYANO) IRON, FE2/S2 (INORGANIC) CLUSTER, FLAVIN MONONUCLEOTIDE, ... | | 著者 | Shomura, Y, Taketa, M, Nakashima, H, Tai, H, Nakagawa, H, Ikeda, Y, Ishii, M, Igarashi, Y, Nishihara, H, Yoon, K.S, Ogo, S, Hirota, S, Higuchi, Y. | | 登録日 | 2017-04-09 | | 公開日 | 2017-08-23 | | 最終更新日 | 2017-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.58 Å) | | 主引用文献 | Structural basis of the redox switches in the NAD(+)-reducing soluble [NiFe]-hydrogenase
Science, 357, 2017
|
|
6NFK
 
 | | Crystal Structure of the Cancer Genomic DNA Mutator APOBEC3B with loop 7 from APOBEC3G bound to iodide | | 分子名称: | 1,2-ETHANEDIOL, DNA dC->dU-editing enzyme APOBEC-3B, IODIDE ION | | 著者 | Shi, K, Orellana, K, Aihara, H. | | 登録日 | 2018-12-20 | | 公開日 | 2019-12-25 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.86 Å) | | 主引用文献 | Active site plasticity and possible modes of chemical inhibition of the human DNA deaminase APOBEC3B
Faseb Bioadv, 2, 2020
|
|
6NFM
 
 | |
6NFL
 
 | | Crystal Structure of the Cancer Genomic DNA Mutator APOBEC3B with loop 7 from APOBEC3G complexed with 2-HP | | 分子名称: | 1,2-ETHANEDIOL, 1,3-diazinan-2-one, CHLORIDE ION, ... | | 著者 | Shi, K, Orellana, K, Aihara, H. | | 登録日 | 2018-12-20 | | 公開日 | 2019-12-25 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.731 Å) | | 主引用文献 | Active site plasticity and possible modes of chemical inhibition of the human DNA deaminase APOBEC3B
Faseb Bioadv, 2, 2020
|
|
1WTJ
 
 | | Crystal Structure of delta1-piperideine-2-carboxylate reductase from Pseudomonas syringae pvar.tomato | | 分子名称: | ureidoglycolate dehydrogenase | | 著者 | Goto, M, Muramatsu, H, Mihara, H, Kurihara, T, Esaki, N, Omi, R, Miyahara, I, Hirotsu, K. | | 登録日 | 2004-11-24 | | 公開日 | 2005-10-04 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Crystal structures of Delta1-piperideine-2-carboxylate/Delta1-pyrroline-2-carboxylate reductase belonging to a new family of NAD(P)H-dependent oxidoreductases: conformational change, substrate recognition, and stereochemistry of the reaction
J.Biol.Chem., 280, 2005
|
|
6X6O
 
 | | Crystal structure of T4 protein Spackle as determined by native SAD phasing | | 分子名称: | CHLORIDE ION, Protein spackle | | 著者 | Shi, K, Kurniawan, F, Banerjee, S, Moeller, N.H, Aihara, H. | | 登録日 | 2020-05-28 | | 公開日 | 2020-09-16 | | 実験手法 | X-RAY DIFFRACTION (1.52 Å) | | 主引用文献 | Crystal structure of bacteriophage T4 Spackle as determined by native SAD phasing.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
6XC1
 
 | | Crystal structure of bacteriophage T4 spackle and lysozyme in orthorhombic form | | 分子名称: | 1,2-ETHANEDIOL, ISOPROPYL ALCOHOL, Lysozyme, ... | | 著者 | Shi, K, Oakland, J.T, Kurniawan, F, Moeller, N.H, Aihara, H. | | 登録日 | 2020-06-07 | | 公開日 | 2020-12-02 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.92 Å) | | 主引用文献 | Structural basis of superinfection exclusion by bacteriophage T4 Spackle.
Commun Biol, 3, 2020
|
|
6P7A
 
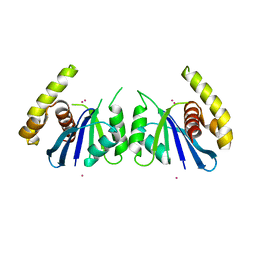 | | CRYSTAL STRUCTURE OF THE FOWLPOX VIRUS HOLLIDAY JUNCTION RESOLVASE | | 分子名称: | CADMIUM ION, Holliday junction resolvase | | 著者 | Li, N, Shi, K, Banerjee, S, Rao, T, Aihara, H. | | 登録日 | 2019-06-05 | | 公開日 | 2020-04-29 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (3.081 Å) | | 主引用文献 | Structural insights into the promiscuous DNA binding and broad substrate selectivity of fowlpox virus resolvase.
Sci Rep, 10, 2020
|
|
8SPH
 
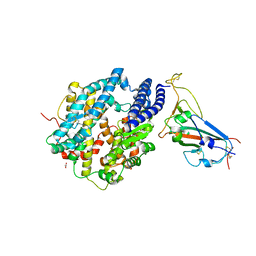 | | Crystal structure of chimeric omicron RBD (strain XBB.1) complexed with human ACE2 | | 分子名称: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Zhang, W, Shi, K, Aihara, H, Li, F. | | 登録日 | 2023-05-03 | | 公開日 | 2023-08-02 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.71 Å) | | 主引用文献 | Structural evolution of SARS-CoV-2 omicron in human receptor recognition.
J.Virol., 97, 2023
|
|
8SPI
 
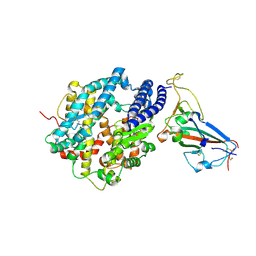 | | Crystal structure of chimeric omicron RBD (strain XBB.1.5) complexed with human ACE2 | | 分子名称: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Zhang, W, Shi, K, Aihara, H, Li, F. | | 登録日 | 2023-05-03 | | 公開日 | 2023-08-02 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (3.06 Å) | | 主引用文献 | Structural evolution of SARS-CoV-2 omicron in human receptor recognition.
J.Virol., 97, 2023
|
|
6XC0
 
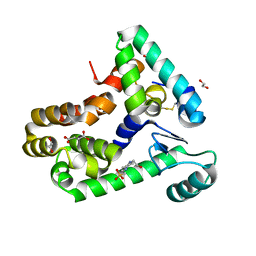 | | Crystal structure of bacteriophage T4 spackle and lysozyme in monoclinic form | | 分子名称: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, ... | | 著者 | Shi, K, Oakland, J.T, Kurniawan, F, Moeller, N.H, Aihara, H. | | 登録日 | 2020-06-07 | | 公開日 | 2020-12-02 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.78 Å) | | 主引用文献 | Structural basis of superinfection exclusion by bacteriophage T4 Spackle.
Commun Biol, 3, 2020
|
|
6P7B
 
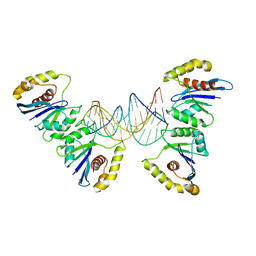 | | Crystal structure of Fowlpox virus resolvase and substrate Holliday junction DNA complex | | 分子名称: | DNA (29-MER), Holliday junction resolvase | | 著者 | Li, N, Shi, K, Rao, T, Banerjee, S, Aihara, H. | | 登録日 | 2019-06-05 | | 公開日 | 2020-04-29 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (3.317 Å) | | 主引用文献 | Structural insights into the promiscuous DNA binding and broad substrate selectivity of fowlpox virus resolvase.
Sci Rep, 10, 2020
|
|
8SBM
 
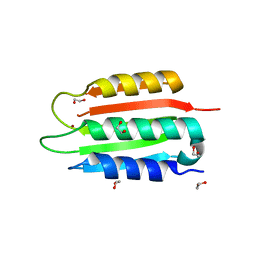 | | Crystal structure of the wild-type Catalytic ATP-binding domain of Mtb DosS | | 分子名称: | 1,2-ETHANEDIOL, GAF domain-containing protein, SODIUM ION, ... | | 著者 | Larson, G, Shi, K, Aihara, H, Bhagi-Damodaran, A. | | 登録日 | 2023-04-03 | | 公開日 | 2023-11-08 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.47 Å) | | 主引用文献 | Understanding ATP Binding to DosS Catalytic Domain with a Short ATP-Lid.
Biochemistry, 62, 2023
|
|
5SXH
 
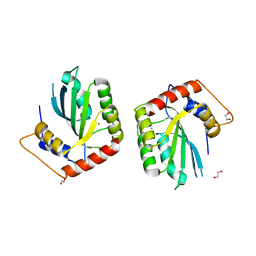 | | Crystal Structure of the Cancer Genomic DNA Mutator APOBEC3B | | 分子名称: | 1,2-ETHANEDIOL, DNA dC->dU-editing enzyme APOBEC-3B, ZINC ION | | 著者 | Shi, K, Kurahashi, K, Aihara, H. | | 登録日 | 2016-08-09 | | 公開日 | 2017-12-27 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.78 Å) | | 主引用文献 | Conformational Switch Regulates the DNA Cytosine Deaminase Activity of Human APOBEC3B.
Sci Rep, 7, 2017
|
|
