2ZJ2
 
 | | Archaeal DNA helicase Hjm apo state in form 1 | | 分子名称: | Putative ski2-type helicase, SULFATE ION | | 著者 | Oyama, T, Oka, H, Fujikane, R, Ishino, Y, Morikawa, K. | | 登録日 | 2008-02-29 | | 公開日 | 2009-02-10 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Atomic structures and functional implications of the archaeal RecQ-like helicase Hjm
Bmc Struct.Biol., 9, 2009
|
|
2ZJ8
 
 | | Archaeal DNA helicase Hjm apo state in form 2 | | 分子名称: | Putative ski2-type helicase | | 著者 | Oyama, T, Oka, H, Fujikane, R, Ishino, Y, Morikawa, K. | | 登録日 | 2008-02-29 | | 公開日 | 2009-02-10 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Atomic structures and functional implications of the archaeal RecQ-like helicase Hjm
Bmc Struct.Biol., 9, 2009
|
|
2ZJ5
 
 | | Archaeal DNA helicase Hjm complexed with ADP in form 1 | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Putative ski2-type helicase, SULFATE ION | | 著者 | Oyama, T, Oka, H, Fujikane, R, Ishino, Y, Morikawa, K. | | 登録日 | 2008-02-29 | | 公開日 | 2009-02-10 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Atomic structures and functional implications of the archaeal RecQ-like helicase Hjm
Bmc Struct.Biol., 9, 2009
|
|
2ZJA
 
 | | Archaeal DNA helicase Hjm complexed with AMPPCP in form 2 | | 分子名称: | PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, Putative ski2-type helicase | | 著者 | Oyama, T, Oka, H, Fujikane, R, Ishino, Y, Morikawa, K. | | 登録日 | 2008-02-29 | | 公開日 | 2009-02-10 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Atomic structures and functional implications of the archaeal RecQ-like helicase Hjm
Bmc Struct.Biol., 9, 2009
|
|
3WV5
 
 | | Complex structure of VinN with 3-methylaspartate | | 分子名称: | (2S,3S)-3-methyl-aspartic acid, Non-ribosomal peptide synthetase | | 著者 | Miyanaga, A, Cieslak, J, Shinohara, Y, Kudo, F, Eguchi, T. | | 登録日 | 2014-05-15 | | 公開日 | 2014-10-01 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | The crystal structure of the adenylation enzyme VinN reveals a unique beta-amino acid recognition mechanism
J.Biol.Chem., 289, 2014
|
|
3WV4
 
 | | Crystal structure of VinN | | 分子名称: | Non-ribosomal peptide synthetase | | 著者 | Miyanaga, A, Cieslak, J, Shinohara, Y, Kudo, F, Eguchi, T. | | 登録日 | 2014-05-15 | | 公開日 | 2014-10-01 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | The crystal structure of the adenylation enzyme VinN reveals a unique beta-amino acid recognition mechanism
J.Biol.Chem., 289, 2014
|
|
3WVN
 
 | | Complex structure of VinN with L-aspartate | | 分子名称: | ASPARTIC ACID, Non-ribosomal peptide synthetase | | 著者 | Miyanaga, A, Cieslak, J, Shinohara, Y, Kudo, F, Eguchi, T. | | 登録日 | 2014-05-30 | | 公開日 | 2014-10-01 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | The crystal structure of the adenylation enzyme VinN reveals a unique beta-amino acid recognition mechanism
J.Biol.Chem., 289, 2014
|
|
6Z1M
 
 | | Structure of an Ancestral glycosidase (family 1) bound to heme | | 分子名称: | 1,2-ETHANEDIOL, Ancestral reconstructed glycosidase, GLYCEROL, ... | | 著者 | Gavira, J.A, Risso, V.A, Sanchez-Ruiz, J.M, Gamiz-Arco, G, Gutierrez-Rus, L, Ibarra-Molero, B, Oshino, Y, Petrovic, D, Romero-Rivera, A, Seelig, B, Kamerlin, S.C.L, Gaucher, E.A. | | 登録日 | 2020-05-14 | | 公開日 | 2020-07-22 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.45 Å) | | 主引用文献 | Heme-binding enables allosteric modulation in an ancient TIM-barrel glycosidase.
Nat Commun, 12, 2021
|
|
9FFD
 
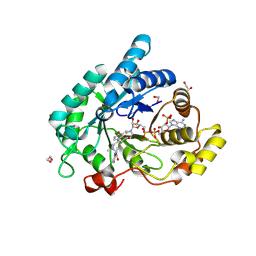 | | STRUCTURE OF ALDO-KETO REDUCTASE 1C3 (AKR1C3) IN COMPLEX WITH AN INHIBITOR MEDS765 | | 分子名称: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Aldo-keto reductase family 1 member C3, ... | | 著者 | Frydenvang, K, Hussain, S, Mirza, O.A. | | 登録日 | 2024-05-23 | | 公開日 | 2024-08-21 | | 最終更新日 | 2024-08-28 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | AI Based Discovery of a New AKR1C3 Inhibitor for Anticancer Applications.
Acs Med.Chem.Lett., 15, 2024
|
|
5O8M
 
 | |
4YOW
 
 | |
4YOU
 
 | |
4YOR
 
 | |
4YOT
 
 | |
4YOV
 
 | |
4YOY
 
 | |
4YOX
 
 | |
7DDM
 
 | | Crystal Structure of PenA39 beta-Lactamase | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, ALA-ALA-ARG-ASP-ALA-ALA-VAL-SER-ASP-ALA-ALA-ALA, ... | | 著者 | Nukaga, M, Hoshino, T, Papp-Wallace, K.M. | | 登録日 | 2020-10-29 | | 公開日 | 2021-11-03 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.2 Å) | | 主引用文献 | Frameshift mutations in genes encoding PBP3 and PBP4 trigger an unusual, extreme beta-lactam resistance phenotype in Burkholderia multivorans
To Be Published
|
|
7VLK
 
 | | eIF2B-SFSV NSs C2-imposed | | 分子名称: | Non-structural protein NS-S, Translation initiation factor eIF-2B subunit alpha, Translation initiation factor eIF-2B subunit beta, ... | | 著者 | Kashiwagi, K, Ito, T. | | 登録日 | 2021-10-04 | | 公開日 | 2021-12-01 | | 最終更新日 | 2024-06-19 | | 実験手法 | ELECTRON MICROSCOPY (2.27 Å) | | 主引用文献 | eIF2B-capturing viral protein NSs suppresses the integrated stress response.
Nat Commun, 12, 2021
|
|
5ZB8
 
 | |
6IRY
 
 | | Crystal structure of the zebrafish cap-specific adenosine methyltransferase bound to SAH | | 分子名称: | 1,2-ETHANEDIOL, PDX1 C-terminal-inhibiting factor 1, S-ADENOSYL-L-HOMOCYSTEINE | | 著者 | Hirano, S, Nishimasu, H, Ishitani, R, Nureki, O. | | 登録日 | 2018-11-14 | | 公開日 | 2018-12-05 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Cap-specific terminal N 6 -methylation of RNA by an RNA polymerase II-associated methyltransferase.
Science, 363, 2019
|
|
6IRX
 
 | |
6IRW
 
 | | Crystal structure of the human cap-specific adenosine methyltransferase bound to SAH | | 分子名称: | Phosphorylated CTD-interacting factor 1, S-ADENOSYL-L-HOMOCYSTEINE | | 著者 | Hirano, S, Nishimasu, H, Ishitani, R, Nureki, O. | | 登録日 | 2018-11-14 | | 公開日 | 2018-12-05 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Cap-specific terminal N 6 -methylation of RNA by an RNA polymerase II-associated methyltransferase.
Science, 363, 2019
|
|
7D6J
 
 | | Human serum albumin complexed with benzbromarone | | 分子名称: | Serum albumin, [3,5-bis(bromanyl)-4-oxidanyl-phenyl]-(2-ethyl-1-benzofuran-3-yl)methanone | | 著者 | Kawai, A, Yamasaki, K. | | 登録日 | 2020-09-30 | | 公開日 | 2021-02-03 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (3.29 Å) | | 主引用文献 | Interaction of Benzbromarone with Subdomains IIIA and IB/IIA on Human Serum Albumin as the Primary and Secondary Binding Regions.
Mol Pharm., 18, 2021
|
|
6IRZ
 
 | | Crystal structure of the zebrafish cap-specific adenosine methyltransferase bound to SAH and m7G-capped RNA | | 分子名称: | 1,2-ETHANEDIOL, 7N-METHYL-8-HYDROGUANOSINE-5'-DIPHOSPHATE, PDX1 C-terminal-inhibiting factor 1, ... | | 著者 | Hirano, S, Nishimasu, H, Ishitani, R, Nureki, O. | | 登録日 | 2018-11-15 | | 公開日 | 2018-12-05 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Cap-specific terminal N 6 -methylation of RNA by an RNA polymerase II-associated methyltransferase.
Science, 363, 2019
|
|
