8DOX
 
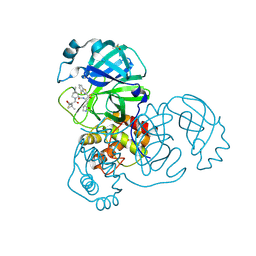 | | Crystal structure of SARS-CoV-2 main protease in complex with an inhibitor TKB-245 | | 分子名称: | (1R,2S,5S)-N-{(1S,2S)-1-(4-fluoro-1,3-benzothiazol-2-yl)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5, DIMETHYL SULFOXIDE | | 著者 | Bulut, H, Hayashi, H, Tsuji, K, Kuwata, N, Das, D, Tamamura, H, Mitsuya, H. | | 登録日 | 2022-07-14 | | 公開日 | 2022-09-21 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.46 Å) | | 主引用文献 | Identification of SARS-CoV-2 M pro inhibitors containing P1' 4-fluorobenzothiazole moiety highly active against SARS-CoV-2.
Nat Commun, 14, 2023
|
|
2Y4W
 
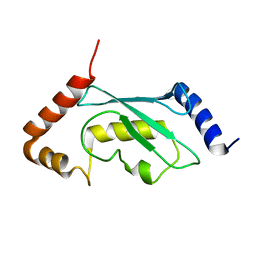 | | Solution structure of human ubiquitin conjugating enzyme Rad6b | | 分子名称: | UBIQUITIN-CONJUGATING ENZYME E2 B | | 著者 | Huang, A, Hibbert, R.G, deJong, R.N, Das, D, Sixma, T.K, Boelens, R. | | 登録日 | 2011-01-11 | | 公開日 | 2011-05-11 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Symmetry and Asymmetry of the Ring-Ring Dimer of Rad18
J.Mol.Biol., 410, 2011
|
|
5COK
 
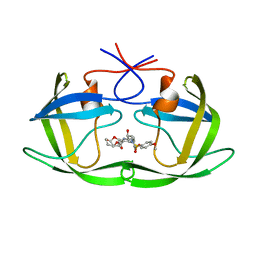 | | X-ray crystal structure of wild type HIV-1 protease in complex with GRL-0476 | | 分子名称: | (3aS,4S,7aR)-hexahydro-4H-furo[2,3-b]pyran-4-yl [(2S,3R)-3-hydroxy-4-{[(4-methoxyphenyl)sulfonyl](2-methylpropyl)amino}-1-phenylbutan-2-yl]carbamate, HIV-1 protease | | 著者 | Yedidi, R.S, Hayashi, H, Aoki, M, Das, D, Ghosh, A.K, Mitsuya, H. | | 登録日 | 2015-07-20 | | 公開日 | 2016-01-13 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.801 Å) | | 主引用文献 | C-5-Modified Tetrahydropyrano-Tetrahydofuran-Derived Protease Inhibitors (PIs) Exert Potent Inhibition of the Replication of HIV-1 Variants Highly Resistant to Various PIs, including Darunavir.
J.Virol., 90, 2015
|
|
5COP
 
 | | X-ray crystal structure of wild type HIV-1 protease in complex with GRL-097 | | 分子名称: | (3R,3aS,4S,7aS)-3-hydroxyhexahydro-4H-furo[2,3-b]pyran-4-yl [(2S,3R)-4-{[(4-aminophenyl)sulfonyl](2-methylpropyl)amino}-3-hydroxy-1-(4-methoxyphenyl)butan-2-yl]carbamate, HIV-1 protease | | 著者 | Yedidi, R.S, Hayashi, H, Aoki, M, Das, D, Ghosh, A.K, Mitsuya, H. | | 登録日 | 2015-07-20 | | 公開日 | 2016-01-13 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | C-5-Modified Tetrahydropyrano-Tetrahydofuran-Derived Protease Inhibitors (PIs) Exert Potent Inhibition of the Replication of HIV-1 Variants Highly Resistant to Various PIs, including Darunavir.
J.Virol., 90, 2015
|
|
5CON
 
 | | X-ray crystal structure of wild type HIV-1 protease in complex with GRL-015 | | 分子名称: | (3R,3aS,4S,7aS)-3-hydroxyhexahydro-4H-furo[2,3-b]pyran-4-yl [(2S,3R)-3-hydroxy-4-{[(4-methoxyphenyl)sulfonyl](2-methylpropyl)amino}-1-phenylbutan-2-yl]carbamate, HIV-1 protease | | 著者 | Yedidi, R.S, Hayashi, H, Aoki, M, Das, D, Ghosh, A.K, Mitsuya, H. | | 登録日 | 2015-07-20 | | 公開日 | 2016-01-13 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | C-5-Modified Tetrahydropyrano-Tetrahydofuran-Derived Protease Inhibitors (PIs) Exert Potent Inhibition of the Replication of HIV-1 Variants Highly Resistant to Various PIs, including Darunavir.
J.Virol., 90, 2015
|
|
5COO
 
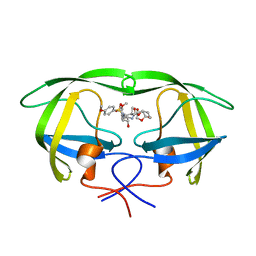 | | X-ray crystal structure of wild type HIV-1 protease in complex with GRL-085 | | 分子名称: | (3R,3aS,4S,7aS)-3-hydroxyhexahydro-4H-furo[2,3-b]pyran-4-yl [(2S,3R)-3-hydroxy-1-(4-methoxyphenyl)-4-{[(4-methoxyphenyl)sulfonyl](2-methylpropyl)amino}butan-2-yl]carbamate, HIV-1 protease | | 著者 | Yedidi, R.S, Hayashi, H, Aoki, M, Das, D, Ghosh, A.K, Mitsuya, H. | | 登録日 | 2015-07-20 | | 公開日 | 2016-01-13 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | C-5-Modified Tetrahydropyrano-Tetrahydofuran-Derived Protease Inhibitors (PIs) Exert Potent Inhibition of the Replication of HIV-1 Variants Highly Resistant to Various PIs, including Darunavir.
J.Virol., 90, 2015
|
|
1NND
 
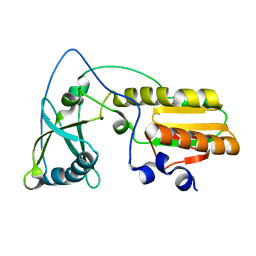 | | Arginine 116 is Essential for Nucleic Acid Recognition by the Fingers Domain of Moloney Murine Leukemia Virus Reverse Transcriptase | | 分子名称: | Reverse Transcriptase | | 著者 | Crowther, R.L, Remeta, D.P, Minetti, C.A, Das, D, Montano, S.P, Georgiadis, M.M. | | 登録日 | 2003-01-13 | | 公開日 | 2004-01-27 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structural and energetic characterization of nucleic acid-binding to the fingers domain of Moloney murine leukemia virus reverse transcriptase
Proteins, 57, 2004
|
|
3H98
 
 | | Crystal structure of HCV NS5b 1b with (1,1-dioxo-2H-[1,2,4]benzothiadiazin-3-yl) azolo[1,5-a]pyrimidine derivative | | 分子名称: | GLYCEROL, N-{3-[5-hydroxy-8-(3-methylbutyl)-7-oxo-7,8-dihydroimidazo[1,2-a]pyrimidin-6-yl]-1,1-dioxido-4H-1,2,4-benzothiadiazin-7-yl}methanesulfonamide, RNA-directed RNA polymerase | | 著者 | Wang, G, Lei, H, Wang, X, Das, D, Mackinnon, C, Montalbetti, C.A.G, Mears, R, Gai, X, Bailey, S, Ruhrmund, D, Hooi, L, Misialek, S, Rajagopalan, R, Cheng, R.K.Y, Barker, J.L, Felicetti, B, Stoycheva, A, Buckman, B, Kossen, K, Seiwert, S, Beigelmana, L. | | 登録日 | 2009-04-30 | | 公開日 | 2009-10-13 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | HCV NS5B polymerase inhibitors 2: Synthesis and in vitro activity of (1,1-dioxo-2H-[1,2,4]benzothiadiazin-3-yl) azolo[1,5-a]pyridine and azolo[1,5-a]pyrimidine derivatives.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
4KVQ
 
 | | Crystal Structure of Prochlorococcus marinus aldehyde-deformylating oxygenase wild type with palmitic acid bound | | 分子名称: | Aldehyde decarbonylase, FE (III) ION, PALMITIC ACID | | 著者 | Levy, C.W, Khara, B, Menon, N, Mansell, D, Das, D, Marsh, E.N.G, Leys, D, Scrutton, N.S. | | 登録日 | 2013-05-23 | | 公開日 | 2013-06-26 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.842 Å) | | 主引用文献 | Production of propane and other short-chain alkanes by structure-based engineering of ligand specificity in aldehyde-deformylating oxygenase.
Chembiochem, 14, 2013
|
|
2JPD
 
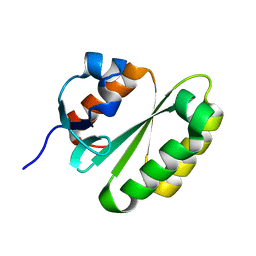 | | Solution structure of the ERCC1 central domain | | 分子名称: | DNA excision repair protein ERCC-1 | | 著者 | Tripsianes, K, Folkers, G, Zheng, C, Das, D, Grinstead, J.S, Kaptein, R, Boelens, R. | | 登録日 | 2007-05-06 | | 公開日 | 2007-09-04 | | 最終更新日 | 2024-05-08 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Analysis of the XPA and ssDNA-binding surfaces on the central domain of human ERCC1 reveals evidence for subfunctionalization
Nucleic Acids Res., 35, 2007
|
|
1Z00
 
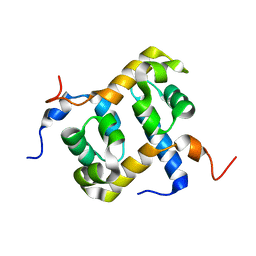 | | Solution structure of the C-terminal domain of ERCC1 complexed with the C-terminal domain of XPF | | 分子名称: | DNA excision repair protein ERCC-1, DNA repair endonuclease XPF | | 著者 | Tripsianes, K, Folkers, G, Ab, E, Das, D, Odijk, H, Jaspers, N.G.J, Hoeijmakers, J.H.J, Kaptein, R, Boelens, R. | | 登録日 | 2005-03-01 | | 公開日 | 2005-12-20 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The Structure of the Human ERCC1/XPF Interaction Domains Reveals a Complementary Role for the Two Proteins in Nucleotide Excision Repair
Structure, 13, 2005
|
|
4NJT
 
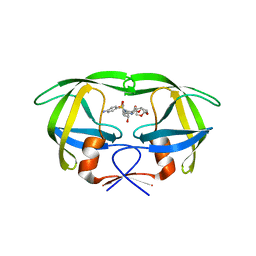 | | Crystal structure of multidrug-resistant clinical isolate A02 HIV-1 protease in complex with darunavir | | 分子名称: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, Protease | | 著者 | Yedidi, R.S, Garimella, H, Chang, S.B, Kaufman, J.D, Das, D, Wingfield, P.T, Ghosh, A.K, Mitsuya, H. | | 登録日 | 2013-11-11 | | 公開日 | 2014-04-02 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | A Conserved Hydrogen-Bonding Network of P2 bis-Tetrahydrofuran-Containing HIV-1 Protease Inhibitors (PIs) with a Protease Active-Site Amino Acid Backbone Aids in Their Activity against PI-Resistant HIV.
Antimicrob.Agents Chemother., 58, 2014
|
|
4NJV
 
 | | Crystal structure of multidrug-resistant clinical isolate A02 HIV-1 protease in complex with ritonavir | | 分子名称: | Protease, RITONAVIR | | 著者 | Yedidi, R.S, Garimella, H, Chang, S.B, Kaufman, J.D, Das, D, Wingfield, P.T, Mitsuya, H. | | 登録日 | 2013-11-11 | | 公開日 | 2014-04-02 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | A Conserved Hydrogen-Bonding Network of P2 bis-Tetrahydrofuran-Containing HIV-1 Protease Inhibitors (PIs) with a Protease Active-Site Amino Acid Backbone Aids in Their Activity against PI-Resistant HIV.
Antimicrob.Agents Chemother., 58, 2014
|
|
4NJS
 
 | | Crystal structure of multidrug-resistant clinical isolate A02 HIV-1 protease in complex with non-peptidic inhibitor, GRL008 | | 分子名称: | (3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl [(2S,3R)-4-{[(4-carbamoylphenyl)sulfonyl](2-methylpropyl)amino}-3-hydroxy-1-phenylbutan-2-yl]carbamate, Protease | | 著者 | Yedidi, R.S, Garimella, H, Kaufman, J.D, Das, D, Wingfield, P.T, Ghosh, A.K, Mitsuya, H. | | 登録日 | 2013-11-11 | | 公開日 | 2014-04-02 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | A Conserved Hydrogen-Bonding Network of P2 bis-Tetrahydrofuran-Containing HIV-1 Protease Inhibitors (PIs) with a Protease Active-Site Amino Acid Backbone Aids in Their Activity against PI-Resistant HIV.
Antimicrob.Agents Chemother., 58, 2014
|
|
4NJU
 
 | | Crystal structure of multidrug-resistant clinical isolate A02 HIV-1 protease in complex with tipranavir | | 分子名称: | N-(3-{(1R)-1-[(6R)-4-HYDROXY-2-OXO-6-PHENETHYL-6-PROPYL-5,6-DIHYDRO-2H-PYRAN-3-YL]PROPYL}PHENYL)-5-(TRIFLUOROMETHYL)-2-PYRIDINESULFONAMIDE, Protease | | 著者 | Yedidi, R.S, Garimella, H, Kaufman, J.D, Das, D, Wingfield, P.T, Mitsuya, H. | | 登録日 | 2013-11-11 | | 公開日 | 2014-04-02 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | A Conserved Hydrogen-Bonding Network of P2 bis-Tetrahydrofuran-Containing HIV-1 Protease Inhibitors (PIs) with a Protease Active-Site Amino Acid Backbone Aids in Their Activity against PI-Resistant HIV.
Antimicrob.Agents Chemother., 58, 2014
|
|
2BJC
 
 | | NMR structure of a protein-DNA complex of an altered specificity mutant of the lac repressor headpiece that mimics the gal repressor | | 分子名称: | 5'-D(*GP*AP*AP*TP*TP*GP*TP*AP*AP*GP *CP*GP*CP*TP*TP*AP*CP*AP*AP*TP*TP*C)-3', LACTOSE OPERON REPRESSOR | | 著者 | Salinas, R.K, Folkers, G.E, Bonvin, A.M.J.J, Das, D, Boelens, R, Kaptein, R. | | 登録日 | 2005-02-01 | | 公開日 | 2005-10-18 | | 最終更新日 | 2020-01-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Altered Specificity in DNA Binding by the Lac Repressor: A Mutant Lac Headpiece that Mimics the Gal Repressor
Chembiochem, 6, 2005
|
|
4KVR
 
 | | Crystal Structure of Prochlorococcus marinus aldehyde-deformylating oxygenase (mutant V41Y) | | 分子名称: | Aldehyde decarbonylase, FE (III) ION, HEXANOIC ACID | | 著者 | Levy, C.W, Khara, B, Menon, N, Mansell, D, Das, D, Marsh, E.N.G, Leys, D, Scrutton, N.S. | | 登録日 | 2013-05-23 | | 公開日 | 2013-06-26 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.88 Å) | | 主引用文献 | Production of propane and other short-chain alkanes by structure-based engineering of ligand specificity in aldehyde-deformylating oxygenase.
Chembiochem, 14, 2013
|
|
4KVS
 
 | | Crystal Structure of Prochlorococcus marinus aldehyde-deformylating oxygenase (mutant A134F) | | 分子名称: | Aldehyde decarbonylase, FE (III) ION, HEXANOIC ACID | | 著者 | Levy, C.W, Khara, B, Menon, N, Mansell, D, Das, D, Marsh, E.N.G, Leys, D, Scrutton, N.S. | | 登録日 | 2013-05-23 | | 公開日 | 2013-06-26 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.67 Å) | | 主引用文献 | Production of propane and other short-chain alkanes by structure-based engineering of ligand specificity in aldehyde-deformylating oxygenase.
Chembiochem, 14, 2013
|
|
4HLA
 
 | | Crystal structure of wild type HIV-1 protease in complex with darunavir | | 分子名称: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, Protease | | 著者 | Yedidi, R.S, Garimella, H, Palmer, I, Das, D, Wingfield, P.T, Ghosh, A.K, Mitsuya, H. | | 登録日 | 2012-10-16 | | 公開日 | 2013-07-24 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | P2' benzene carboxylic acid moiety is associated with decrease in cellular uptake: evaluation of novel non-peptidic HIV-1 protease inhibitors containing P2 bis-tetrahydrofuran moiety.
Antimicrob.Agents Chemother., 57, 2013
|
|
4I8W
 
 | | Crystal structure of wild type HIV-1 protease in complex with non-peptidic inhibitor, GRL007 | | 分子名称: | 4-{[(2R,3S)-3-({[(3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yloxy]carbonyl}amino)-2-hydroxy-4-phenylbutyl](2-methylpropyl)sulfamoyl}benzoic acid, Protease | | 著者 | Yedidi, R.S, Palmer, I, Das, D, Wingfield, P.T, Ghosh, A.K, Mitsuya, H. | | 登録日 | 2012-12-04 | | 公開日 | 2013-07-24 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.96 Å) | | 主引用文献 | P2' benzene carboxylic acid moiety is associated with decrease in cellular uptake: evaluation of novel non-peptidic HIV-1 protease inhibitors containing P2 bis-tetrahydrofuran moiety.
Antimicrob.Agents Chemother., 57, 2013
|
|
4I8Z
 
 | | Crystal structure of wild type HIV-1 protease in complex with non-peptidic inhibitor, GRL008 | | 分子名称: | (3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl [(2S,3R)-4-{[(4-carbamoylphenyl)sulfonyl](2-methylpropyl)amino}-3-hydroxy-1-phenylbutan-2-yl]carbamate, Protease | | 著者 | Yedidi, R.S, Palmer, I, Das, D, Wingfield, P.T, Ghosh, A.K, Mitsuya, H. | | 登録日 | 2012-12-04 | | 公開日 | 2013-07-24 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | P2' benzene carboxylic acid moiety is associated with decrease in cellular uptake: evaluation of novel non-peptidic HIV-1 protease inhibitors containing P2 bis-tetrahydrofuran moiety.
Antimicrob.Agents Chemother., 57, 2013
|
|
3EOV
 
 | | Crystal structure of cyclophilin from Leishmania donovani ligated with cyclosporin A | | 分子名称: | CYCLOSPORIN A, PEPTIDYL-PROLYL CIS-TRANS ISOMERASE | | 著者 | Venugopal, V, Dasgupta, D, Datta, A.K, Banerjee, R. | | 登録日 | 2008-09-29 | | 公開日 | 2008-11-11 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Structure of Cyclophilin from Leishmania Donovani Bound to Cyclosporin at 2.6 A Resolution: Correlation between Structure and Thermodynamic Data.
Acta Crystallogr.,Sect.D, 65, 2009
|
|
2YKT
 
 | | Crystal structure of the I-BAR domain of IRSp53 (BAIAP2) in complex with an EHEC derived Tir peptide | | 分子名称: | BRAIN-SPECIFIC ANGIOGENESIS INHIBITOR 1-ASSOCIATED PROTEIN 2, SULFATE ION, TRANSLOCATED INTIMIN RECEPTOR PROTEIN | | 著者 | de Groot, J.C, Schlueter, K, Carius, Y, Quedenau, C, Vingadassalom, D, Faix, J, Weiss, S.M, Reichelt, J, Standfuss-Gabisch, C, Lesser, C.F, Leong, J.M, Heinz, D.W, Buessow, K, Stradal, T.E.B. | | 登録日 | 2011-05-30 | | 公開日 | 2011-09-07 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.11 Å) | | 主引用文献 | Structural Basis for Complex Formation between Human Irsp53 and the Translocated Intimin Receptor Tir of Enterohemorrhagic E. Coli.
Structure, 19, 2011
|
|
3DEE
 
 | |
3K5J
 
 | |
