3LML
 
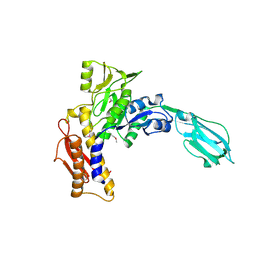 | | Crystal Structure of the sheath tail protein Lin1278 from Listeria innocua, Northeast Structural Genomics Consortium Target LkR115 | | 分子名称: | Lin1278 protein | | 著者 | Forouhar, F, Neely, H, Seetharaman, J, Mao, M, Xiao, R, Patel, D.J, Ciccosanti, C, Wang, H, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2010-01-31 | | 公開日 | 2010-03-09 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (3.3 Å) | | 主引用文献 | Northeast Structural Genomics Consortium Target LkR115
To be Published
|
|
3LQI
 
 | |
3LQJ
 
 | |
6IEJ
 
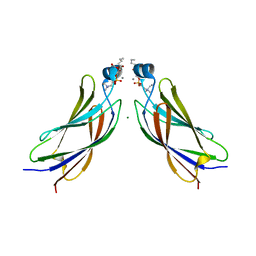 | | The C2 domain of cytosolic phospholipase A2 alpha bound to phosphatidylcholine | | 分子名称: | 1,2-dihexanoyl-sn-glycero-3-phosphocholine, CALCIUM ION, Cytosolic phospholipase A2, ... | | 著者 | Hirano, Y, Gao, Y.G, Stephenson, D.J, Vu, N.T, Malinina, L, Chalfant, C.E, Patel, D.J, Brown, R.E. | | 登録日 | 2018-09-14 | | 公開日 | 2019-05-22 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.206 Å) | | 主引用文献 | Structural basis of phosphatidylcholine recognition by the C2-domain of cytosolic phospholipase A2alpha.
Elife, 8, 2019
|
|
4FSX
 
 | |
4FT4
 
 | |
5KDI
 
 | | How FAPP2 Selects Simple Glycosphingolipids Using the GLTP-fold | | 分子名称: | (~{Z})-~{N}-[(~{E},2~{S},3~{R})-1-[(2~{R},3~{R},4~{S},5~{R},6~{R})-6-(hydroxymethyl)-3,4,5-tris(oxidanyl)oxan-2-yl]oxy-3-oxidanyl-octadec-4-en-2-yl]octadec-9-enamide, Pleckstrin homology domain-containing family A member 8 | | 著者 | Ochoa-Lizarralde, B, Popov, A.N, Samygina, V.R, Patel, D.J, Brown, R.E, Malinina, L. | | 登録日 | 2016-06-08 | | 公開日 | 2017-12-13 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | Structural analyses of 4-phosphate adaptor protein 2 yield mechanistic insights into sphingolipid recognition by the glycolipid transfer protein family.
J.Biol.Chem., 293, 2018
|
|
5KK5
 
 | | AsCpf1(E993A)-crRNA-DNA ternary complex | | 分子名称: | CRISPR-associated endonuclease Cpf1, DNA (28-MER), DNA (8-mer), ... | | 著者 | Gao, P, Yang, H, Rajashankar, K.R, Huang, Z, Patel, D.J. | | 登録日 | 2016-06-21 | | 公開日 | 2016-08-10 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (3.289 Å) | | 主引用文献 | Type V CRISPR-Cas Cpf1 endonuclease employs a unique mechanism for crRNA-mediated target DNA recognition.
Cell Res., 26, 2016
|
|
6D92
 
 | | Ternary RsAgo Complex with Guide RNA and Target DNA Containing A-A non-canonical pair at position 3 | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, DNA (5'-D(P*TP*CP*GP*TP*CP*AP*CP*CP*TP*GP*TP*GP*CP*AP*GP*AP*AP*AP*C)-3'), ... | | 著者 | Liu, Y, Esyunina, D, Olovnikov, I, Teplova, M, Patel, D.J. | | 登録日 | 2018-04-27 | | 公開日 | 2018-07-25 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.81 Å) | | 主引用文献 | Accommodation of Helical Imperfections in Rhodobacter sphaeroides Argonaute Ternary Complexes with Guide RNA and Target DNA.
Cell Rep, 24, 2018
|
|
6D8F
 
 | | RsAgo Ternary Complex with Guide RNA and Target DNA Containing T-T Bulge Within the Seed Segment | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, DNA (5'-D(P*TP*CP*GP*TP*CP*AP*CP*CP*TP*GP*TP*GP*CP*AP*GP*TP*TP*TP*AP*AP*C)-3'), ... | | 著者 | Liu, Y, Esyunina, D, Olovnikov, I, Teplova, M, Patel, D.J. | | 登録日 | 2018-04-26 | | 公開日 | 2018-07-25 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | Accommodation of Helical Imperfections in Rhodobacter sphaeroides Argonaute Ternary Complexes with Guide RNA and Target DNA.
Cell Rep, 24, 2018
|
|
6D8A
 
 | | RsAgo Ternary Complex with guide RNA and Target DNA Containing A-A Bulge Within the Seed Segment of the Target Strand | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, DNA (5'-D(P*TP*CP*GP*TP*CP*AP*CP*CP*TP*GP*TP*GP*CP*AP*GP*AP*AP*TP*AP*AP*C)-3'), ... | | 著者 | Liu, Y, Esyunina, D, Olovnikov, I, Teplova, M, Patel, D.J. | | 登録日 | 2018-04-26 | | 公開日 | 2018-07-25 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.251 Å) | | 主引用文献 | Accommodation of Helical Imperfections in Rhodobacter sphaeroides Argonaute Ternary Complexes with Guide RNA and Target DNA.
Cell Rep, 24, 2018
|
|
9C67
 
 | |
9C69
 
 | |
9C6C
 
 | |
9CDB
 
 | | CryoEM structure of CRISPR associated effector, CARF-Adenosine deaminase 1, Cad1, in cA6 (partial density) bound form with ATP (partial density). | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, Adenosine deaminase domain-containing protein, MAGNESIUM ION, ... | | 著者 | Majumder, P, Patel, D.J. | | 登録日 | 2024-06-24 | | 公開日 | 2024-10-30 | | 最終更新日 | 2024-11-06 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | The CRISPR-associated adenosine deaminase Cad1 converts ATP to ITP to provide antiviral immunity.
Cell, 2024
|
|
9C68
 
 | |
9C6A
 
 | |
9C77
 
 | | cryoEM structure of CRISPR associated effector, CARF-Adenosine deaminase 1, Cad1, in cA4 bound form with ATP. | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, Adenosine deaminase domain-containing protein, MAGNESIUM ION, ... | | 著者 | Majumder, P, Patel, D.J. | | 登録日 | 2024-06-10 | | 公開日 | 2024-10-30 | | 最終更新日 | 2024-11-06 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | The CRISPR-associated adenosine deaminase Cad1 converts ATP to ITP to provide antiviral immunity.
Cell, 2024
|
|
9C6F
 
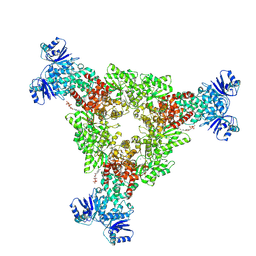 | |
7DMQ
 
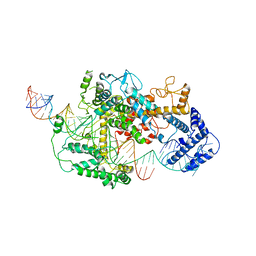 | | Cryo-EM structure of LshCas13a-crRNA-anti-tag RNA complex | | 分子名称: | Anti-tag target RNA, CRISPR RNA, CRISPR/Cas system Cas13a | | 著者 | Wang, B, Zhang, T, Ding, J, Patel, D.J, Yang, H. | | 登録日 | 2020-12-05 | | 公開日 | 2021-02-10 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.06 Å) | | 主引用文献 | Structural basis for self-cleavage prevention by tag:anti-tag pairing complementarity in type VI Cas13 CRISPR systems.
Mol.Cell, 81, 2021
|
|
4NJ5
 
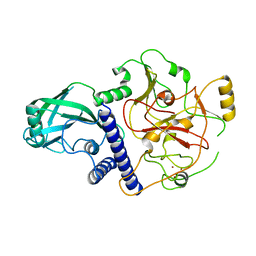 | | Crystal structure of SUVH9 | | 分子名称: | Probable histone-lysine N-methyltransferase, H3 lysine-9 specific SUVH9, ZINC ION | | 著者 | Du, J, Patel, D.J. | | 登録日 | 2013-11-08 | | 公開日 | 2014-01-22 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | SRA- and SET-domain-containing proteins link RNA polymerase V occupancy to DNA methylation.
Nature, 507, 2014
|
|
1D6D
 
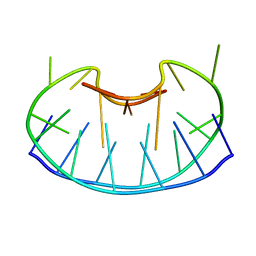 | | SOLUTION DNA STRUCTURE CONTAINING (A-A)-T TRIADS INTERDIGITATED BETWEEN A-T BASE PAIRS AND GGGG TETRADS; NMR, 8 STRUCT. | | 分子名称: | 5'-D(*AP*AP*GP*GP*TP*TP*TP*TP*AP*AP*GP*G)-3' | | 著者 | Kuryavyi, V.V, Kettani, A, Wang, W, Jones, R, Patel, D.J. | | 登録日 | 1999-10-13 | | 公開日 | 2000-01-17 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | A diamond-shaped zipper-like DNA architecture containing triads sandwiched between mismatches and tetrads.
J.Mol.Biol., 295, 2000
|
|
1CR3
 
 | | SOLUTION CONFORMATION OF THE (+)TRANS-ANTI-BENZO[G]CHRYSENE-DA ADDUCT OPPOSITE DT IN A DNA DUPLEX | | 分子名称: | BENZO[G]CHRYSENE, DNA (5'-D(*CP*TP*CP*TP*CP*AP*CP*TP*TP*CP*C)-3'), DNA (5'-D(*GP*GP*AP*AP*GP*TP*GP*AP*GP*AP*G)-3') | | 著者 | Suri, A.K, Mao, B, Amin, S, Geacintov, N.E, Patel, D.J. | | 登録日 | 1999-08-12 | | 公開日 | 2000-02-18 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution conformation of the (+)-trans-anti-benzo[g]chrysene-dA adduct opposite dT in a DNA duplex.
J.Mol.Biol., 292, 1999
|
|
1D83
 
 | | STRUCTURE REFINEMENT OF THE CHROMOMYCIN DIMER/DNA OLIGOMER COMPLEX IN SOLUTION | | 分子名称: | (1S)-5-deoxy-1-O-methyl-1-C-[(2R,3S)-3,5,7,10-tetrahydroxy-6-methyl-4-oxo-1,2,3,4-tetrahydroanthracen-2-yl]-D-xylulose, 2,6-dideoxy-4-O-methyl-alpha-D-galactopyranose-(1-3)-(2R,3R,6R)-6-hydroxy-2-methyltetrahydro-2H-pyran-3-yl acetate, 3-C-methyl-4-O-acetyl-alpha-L-Olivopyranose-(1-3)-(2R,5S,6R)-6-methyltetrahydro-2H-pyran-2,5-diol-(1-3)-(2R,5S,6R)-6-methyltetrahydro-2H-pyran-2,5-diol, ... | | 著者 | Gao, X, Mirau, P, Patel, D.J. | | 登録日 | 1992-07-22 | | 公開日 | 1993-07-15 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structure refinement of the chromomycin dimer-DNA oligomer complex in solution.
J.Mol.Biol., 223, 1992
|
|
7K3K
 
 | |
