7EPD
 
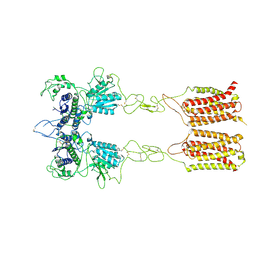 | | Cryo-EM structure of inactive mGlu2-7 heterodimer | | 分子名称: | Isoform 3 of Metabotropic glutamate receptor 7, Metabotropic glutamate receptor 2,Peptidylprolyl isomerase | | 著者 | Du, J, Wang, D, Fan, H, Tai, L, Lin, S, Han, S, Sun, F, Wu, B, Zhao, Q. | | 登録日 | 2021-04-26 | | 公開日 | 2021-06-23 | | 最終更新日 | 2021-07-07 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | Structures of human mGlu2 and mGlu7 homo- and heterodimers.
Nature, 594, 2021
|
|
7EPA
 
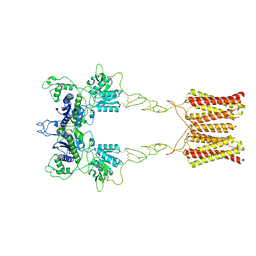 | | Cryo-EM structure of inactive mGlu2 homodimer | | 分子名称: | Metabotropic glutamate receptor 2 | | 著者 | Du, J, Wang, D, Fan, H, Tai, L, Lin, S, Han, S, Sun, F, Wu, B, Zhao, Q. | | 登録日 | 2021-04-26 | | 公開日 | 2021-06-23 | | 最終更新日 | 2021-07-07 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Structures of human mGlu2 and mGlu7 homo- and heterodimers.
Nature, 594, 2021
|
|
7EPE
 
 | | Crystal structure of mGlu2 bound to NAM563 | | 分子名称: | 4-(1-methylpyrazol-4-yl)-7-[[(2~{S})-2-(trifluoromethyl)morpholin-4-yl]methyl]quinoline-2-carboxamide, FLAVIN MONONUCLEOTIDE, Metabotropic glutamate receptor 2 | | 著者 | Du, J, Wang, D, Lin, S, Han, S, Wu, B, Zhao, Q. | | 登録日 | 2021-04-26 | | 公開日 | 2021-06-23 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structures of human mGlu2 and mGlu7 homo- and heterodimers.
Nature, 594, 2021
|
|
7EPF
 
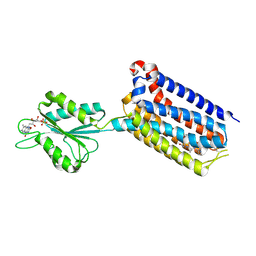 | | Crystal structure of mGlu2 bound to NAM597 | | 分子名称: | (8~{R})-4-[2,4-bis(fluoranyl)phenyl]-8-methyl-7-[(2-methylpyrazol-3-yl)methyl]-6,8-dihydro-5~{H}-1,7-naphthyridine-2-carboxamide, FLAVIN MONONUCLEOTIDE, Metabotropic glutamate receptor 2 | | 著者 | Du, J, Wang, D, Lin, S, Han, S, Wu, B, Zhao, Q. | | 登録日 | 2021-04-26 | | 公開日 | 2021-06-23 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Structures of human mGlu2 and mGlu7 homo- and heterodimers.
Nature, 594, 2021
|
|
6MEO
 
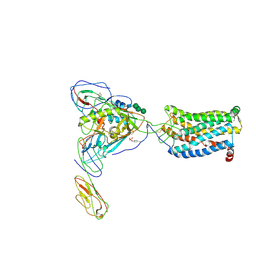 | | Structural basis of coreceptor recognition by HIV-1 envelope spike | | 分子名称: | 2-acetamido-2-deoxy-alpha-D-galactopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Shaik, M.M, Chen, B. | | 登録日 | 2018-09-06 | | 公開日 | 2018-12-12 | | 最終更新日 | 2020-07-29 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | Structural basis of coreceptor recognition by HIV-1 envelope spike.
Nature, 565, 2018
|
|
7WK8
 
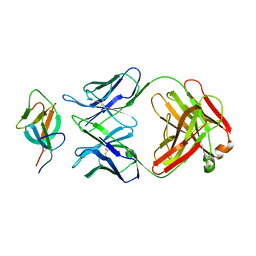 | |
7WKA
 
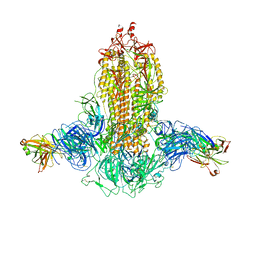 | |
7WK9
 
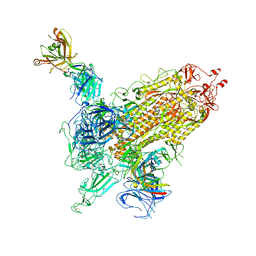 | |
7WK2
 
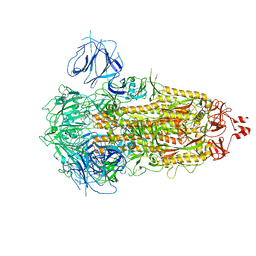 | | SARS-CoV-2 Omicron S-close | | 分子名称: | Spike glycoprotein | | 著者 | Li, J.W, Cong, Y. | | 登録日 | 2022-01-08 | | 公開日 | 2022-01-26 | | 最終更新日 | 2022-05-04 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Molecular basis of receptor binding and antibody neutralization of Omicron.
Nature, 604, 2022
|
|
7WK6
 
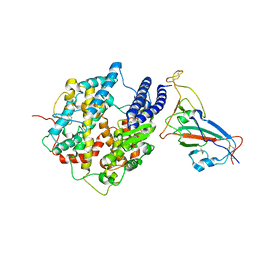 | |
7WK4
 
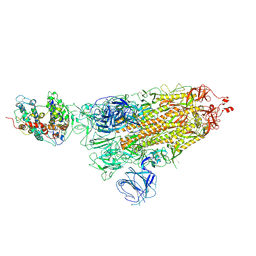 | |
7WVO
 
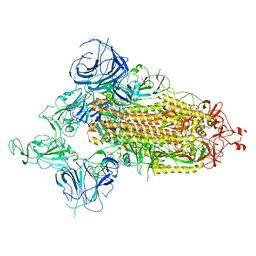 | | SARS-CoV-2 Omicron S-open-2 | | 分子名称: | Spike glycoprotein | | 著者 | Li, J.W, Cong, Y. | | 登録日 | 2022-02-10 | | 公開日 | 2022-03-02 | | 最終更新日 | 2022-05-04 | | 実験手法 | ELECTRON MICROSCOPY (3.41 Å) | | 主引用文献 | Molecular basis of receptor binding and antibody neutralization of Omicron.
Nature, 604, 2022
|
|
7WVN
 
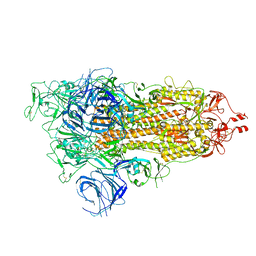 | | SARS-CoV-2 Omicron S-open | | 分子名称: | Spike glycoprotein | | 著者 | Li, J.W, Cong, Y. | | 登録日 | 2022-02-10 | | 公開日 | 2022-03-02 | | 最終更新日 | 2022-05-04 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Molecular basis of receptor binding and antibody neutralization of Omicron.
Nature, 604, 2022
|
|
7ZUD
 
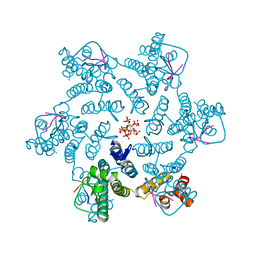 | | Crystal structure of HIV-1 capsid IP6-CPSF6 complex | | 分子名称: | Capsid protein p24, Cleavage and polyadenylation specificity factor subunit 6, INOSITOL HEXAKISPHOSPHATE | | 著者 | Nicastro, G, Taylor, I.A. | | 登録日 | 2022-05-12 | | 公開日 | 2022-07-27 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.93 Å) | | 主引用文献 | CP-MAS and Solution NMR Studies of Allosteric Communication in CA-assemblies of HIV-1.
J.Mol.Biol., 434, 2022
|
|
4D8L
 
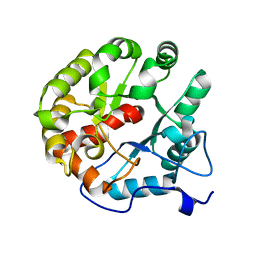 | | Crystal structure of the 2-pyrone-4,6-dicarboxylic acid hydrolase from sphingomonas paucimobilis | | 分子名称: | 2-pyrone-4,6-dicarbaxylate hydrolase | | 著者 | Malashkevich, V.N, Toro, R, Bonanno, J, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | 登録日 | 2012-01-10 | | 公開日 | 2012-01-25 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structure and Catalytic Mechanism of LigI: Insight into the Amidohydrolase Enzymes of cog3618 and Lignin Degradation.
Biochemistry, 51, 2012
|
|
6JLU
 
 | | Structure of PSII-FCP supercomplex from a centric diatom Chaetoceros gracilis at 3.02 angstrom resolution | | 分子名称: | (3S,3'R,5R,6S,7cis)-7',8'-didehydro-5,6-dihydro-5,6-epoxy-beta,beta-carotene-3,3'-diol, (3S,3'S,5R,5'R,6S,6'R,8'R)-3,5'-dihydroxy-8-oxo-6',7'-didehydro-5,5',6,6',7,8-hexahydro-5,6-epoxy-beta,beta-caroten-3'-yl acetate, 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, ... | | 著者 | Pi, X, Zhao, S, Wang, W, Kuang, T, Sui, S, Shen, J. | | 登録日 | 2019-03-06 | | 公開日 | 2019-07-31 | | 最終更新日 | 2019-11-06 | | 実験手法 | ELECTRON MICROSCOPY (3.02 Å) | | 主引用文献 | The pigment-protein network of a diatom photosystem II-light-harvesting antenna supercomplex.
Science, 365, 2019
|
|
4DI8
 
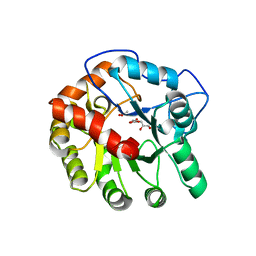 | | CRYSTAL STRUCTURE OF THE D248A mutant of 2-PYRONE-4,6-DICARBOXYLIC ACID HYDROLASE FROM SPHINGOMONAS PAUCIMOBILIS complexed with substrate at pH 8.5 | | 分子名称: | (1E,3Z)-4-hydroxybuta-1,3-diene-1,2,4-tricarboxylic acid, 2-oxo-2H-pyran-4,6-dicarboxylic acid, 2-pyrone-4,6-dicarbaxylate hydrolase, ... | | 著者 | Malashkevich, V.N, Toro, R, Hobbs, M.E, Raushel, F.M, Almo, S.C. | | 登録日 | 2012-01-11 | | 公開日 | 2012-10-03 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.81 Å) | | 主引用文献 | Structure and Catalytic Mechanism of LigI: Insight into the Amidohydrolase Enzymes of cog3618 and Lignin Degradation.
Biochemistry, 51, 2012
|
|
4DI9
 
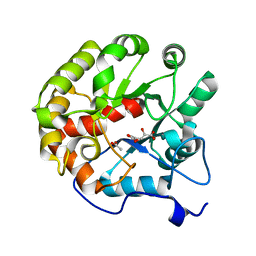 | | CRYSTAL STRUCTURE OF THE D248A mutant of 2-PYRONE-4,6-DICARBOXYLIC ACID HYDROLASE FROM SPHINGOMONAS PAUCIMOBILIS complexed with substrate at pH 6.5 | | 分子名称: | (1E,3Z)-4-hydroxybuta-1,3-diene-1,2,4-tricarboxylic acid, 2-pyrone-4,6-dicarbaxylate hydrolase, ACETATE ION | | 著者 | Malashkevich, V.N, Toro, R, Hobbs, M.E, Raushel, F.M, Almo, S.C. | | 登録日 | 2012-01-11 | | 公開日 | 2012-10-03 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.35 Å) | | 主引用文献 | Structure and Catalytic Mechanism of LigI: Insight into the Amidohydrolase Enzymes of cog3618 and Lignin Degradation.
Biochemistry, 51, 2012
|
|
4DIA
 
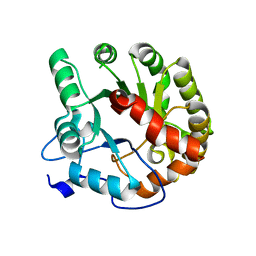 | | CRYSTAL STRUCTURE OF THE D248N mutant of 2-PYRONE-4,6-DICARBOXYLIC ACID HYDROLASE FROM SPHINGOMONAS PAUCIMOBILIS complexed with substrate at pH 4.6 | | 分子名称: | 2-pyrone-4,6-dicarbaxylate hydrolase | | 著者 | Malashkevich, V.N, Toro, R, Hobbs, M.E, Raushel, F.M, Almo, S.C. | | 登録日 | 2012-01-11 | | 公開日 | 2012-10-03 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structure and Catalytic Mechanism of LigI: Insight into the Amidohydrolase Enzymes of cog3618 and Lignin Degradation.
Biochemistry, 51, 2012
|
|
3HK5
 
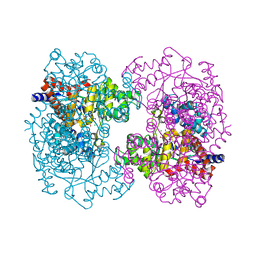 | | Crystal structure of uronate isomerase from Bacillus halodurans complexed with zinc and D-Arabinarate | | 分子名称: | CARBONATE ION, CHLORIDE ION, D-arabinaric acid, ... | | 著者 | Fedorov, A.A, Fedorov, E.V, Nguyen, T.T, Raushel, F.M, Almo, S.C. | | 登録日 | 2009-05-22 | | 公開日 | 2009-08-25 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | The mechanism of the reaction catalyzed by uronate isomerase illustrates how an isomerase may have evolved from a hydrolase within the amidohydrolase superfamily.
Biochemistry, 48, 2009
|
|
3HK8
 
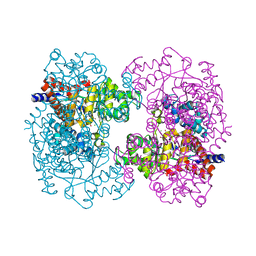 | | Crystal structure of uronate isomerase from Bacillus halodurans complexed with zinc and D-Arabinohydroxamate | | 分子名称: | CARBONATE ION, CHLORIDE ION, D-arabinohydroxamic acid, ... | | 著者 | Fedorov, A.A, Fedorov, E.V, Nguyen, T.T, Raushel, F.M, Almo, S.C. | | 登録日 | 2009-05-22 | | 公開日 | 2009-08-25 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | The mechanism of the reaction catalyzed by uronate isomerase illustrates how an isomerase may have evolved from a hydrolase within the amidohydrolase superfamily.
Biochemistry, 48, 2009
|
|
3HKA
 
 | | Crystal structure of uronate isomerase from Bacillus halodurans complexed with zinc and D-Fructuronate | | 分子名称: | CARBONATE ION, CHLORIDE ION, D-fructuronic acid, ... | | 著者 | Fedorov, A.A, Fedorov, E.V, Nguyen, T.T, Raushel, F.M, Almo, S.C. | | 登録日 | 2009-05-22 | | 公開日 | 2009-08-25 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | The mechanism of the reaction catalyzed by uronate isomerase illustrates how an isomerase may have evolved from a hydrolase within the amidohydrolase superfamily.
Biochemistry, 48, 2009
|
|
6BHR
 
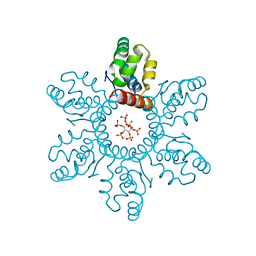 | | HIV-1 immature CTD-SP1 hexamer in complex with IP6 | | 分子名称: | Capsid protein p24,Spacer peptide 1, INOSITOL HEXAKISPHOSPHATE | | 著者 | Zadrozny, K, Wagner, J.M, Ganser-Pornillos, B.K, Pornillos, O. | | 登録日 | 2017-10-31 | | 公開日 | 2018-08-08 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.908 Å) | | 主引用文献 | Inositol phosphates are assembly co-factors for HIV-1.
Nature, 560, 2018
|
|
6BHT
 
 | | HIV-1 CA hexamer in complex with IP6, orthorhombic crystal form | | 分子名称: | Capsid protein p24, INOSITOL HEXAKISPHOSPHATE | | 著者 | Zadrozny, K, Wagner, J.M, Ganser-Pornillos, B.K, Pornillos, O. | | 登録日 | 2017-10-31 | | 公開日 | 2018-08-01 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.689 Å) | | 主引用文献 | Inositol phosphates are assembly co-factors for HIV-1.
Nature, 560, 2018
|
|
6BHS
 
 | | HIV-1 CA hexamer in complex with IP6, hexagonal crystal form | | 分子名称: | Capsid protein p24, INOSITOL HEXAKISPHOSPHATE | | 著者 | Zadrozny, K, Wagner, J.M, Ganser-Pornillos, B.K, Pornillos, O. | | 登録日 | 2017-10-31 | | 公開日 | 2018-08-01 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.984 Å) | | 主引用文献 | Inositol phosphates are assembly co-factors for HIV-1.
Nature, 560, 2018
|
|
