7DAN
 
 | |
7D4Y
 
 | |
7D5R
 
 | | Structure of the Ca2+-bound C646A mutant of peptidylarginine deiminase type III (PAD3) | | 分子名称: | CALCIUM ION, CHLORIDE ION, GLYCEROL, ... | | 著者 | Mashimo, R, Akimoto, M, Unno, M. | | 登録日 | 2020-09-28 | | 公開日 | 2021-06-02 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (3.148 Å) | | 主引用文献 | Structures of human peptidylarginine deiminase type III provide insights into substrate recognition and inhibitor design.
Arch.Biochem.Biophys., 708, 2021
|
|
7D8N
 
 | | Structure of the inactive form of wild-type peptidylarginine deiminase type III (PAD3) crystallized under the condition with high concentrations of Ca2+ | | 分子名称: | CALCIUM ION, CHLORIDE ION, GLYCEROL, ... | | 著者 | Funabashi, K, Sawata, M, Unno, M. | | 登録日 | 2020-10-08 | | 公開日 | 2021-06-02 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.753 Å) | | 主引用文献 | Structures of human peptidylarginine deiminase type III provide insights into substrate recognition and inhibitor design.
Arch.Biochem.Biophys., 708, 2021
|
|
5AVJ
 
 | |
5AVK
 
 | |
7CTQ
 
 | | Peptidyl tryptophan dihydroxylase QhpG essential for tryptophylquinone cofactor biogenesis | | 分子名称: | (2~{R},3~{R},4~{S},5~{S},6~{R})-2-[(2~{R},3~{S},4~{R},5~{R},6~{R})-6-(cyclohexylmethoxy)-2-(hydroxymethyl)-4,5-bis(oxidanyl)oxan-3-yl]oxy-6-(hydroxymethyl)oxane-3,4,5-triol, FLAVIN-ADENINE DINUCLEOTIDE, HEXANE-1,6-DIOL, ... | | 著者 | Oozeki, T, Nakai, T, Okajima, T. | | 登録日 | 2020-08-20 | | 公開日 | 2021-02-17 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (1.978 Å) | | 主引用文献 | Functional and structural characterization of a flavoprotein monooxygenase essential for biogenesis of tryptophylquinone cofactor.
Nat Commun, 12, 2021
|
|
9EN6
 
 | | Crystal structure of RNA G2C4 repeats - native model pH 6.5 | | 分子名称: | MAGNESIUM ION, RNA (5'-R(*GP*GP*CP*CP*CP*C)-3') | | 著者 | Mateja-Pluta, M, Kiliszek, A. | | 登録日 | 2024-03-12 | | 公開日 | 2024-05-01 | | 最終更新日 | 2024-07-03 | | 実験手法 | X-RAY DIFFRACTION (0.918 Å) | | 主引用文献 | Antisense RNA C9orf72 hexanucleotide repeat associated with amyotrophic lateral sclerosis and frontotemporal dementia forms a triplex-like structure and binds small synthetic ligand.
Nucleic Acids Res., 52, 2024
|
|
6L93
 
 | |
6LF1
 
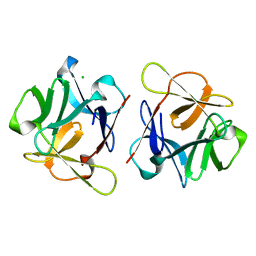 | | SeviL, a GM1b/asialo-GM1 binding lectin | | 分子名称: | CHLORIDE ION, SeviL | | 著者 | Kamata, K, Ozeki, Y, Park, S.-Y, Tame, J.R.H. | | 登録日 | 2019-11-28 | | 公開日 | 2020-12-02 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | The structure of SeviL, a GM1b/asialo-GM1 binding R-type lectin from the mussel Mytilisepta virgata.
Sci Rep, 10, 2020
|
|
6LF2
 
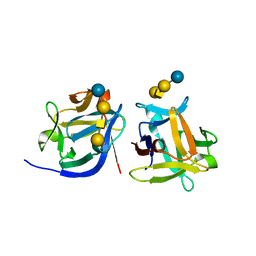 | | SeviL bound to asialo-GM1 saccharide | | 分子名称: | SeviL, beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-galactopyranose-(1-4)-beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | 著者 | Kamata, K, Ozeki, Y, Park, S.-Y, Tame, J.R.H. | | 登録日 | 2019-11-28 | | 公開日 | 2020-12-02 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | The structure of SeviL, a GM1b/asialo-GM1 binding R-type lectin from the mussel Mytilisepta virgata.
Sci Rep, 10, 2020
|
|
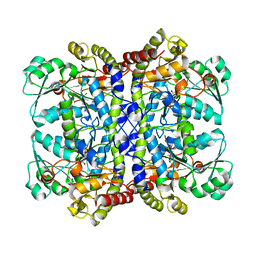
5X2V
 
 | |
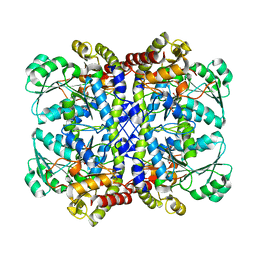
5X2Y
 
 | |
5X2W
 
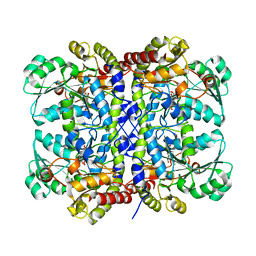 | |
5X2Z
 
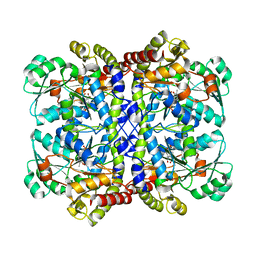 | |
5X30
 
 | | Crystal structure of Pseudomonas putida methionine gamma-lyase C116H mutant with L-homocysteine intermediates. | | 分子名称: | (2E)-2-{[(1E)-{3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene]amino}but-2-enoic acid, (2~{S})-2-[[2-methyl-3-oxidanyl-5-(phosphonooxymethyl)pyridin-4-yl]methylamino]-4-sulfanyl-butanoic acid, 2-AMINO-4-MERCAPTO-BUTYRIC ACID, ... | | 著者 | Shiba, T, Sato, D, Harada, S. | | 登録日 | 2017-02-02 | | 公開日 | 2017-04-12 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structural and mechanistic insights into homocysteine degradation by a mutant of methionine gamma-lyase based on substrate-assisted catalysis
Protein Sci., 26, 2017
|
|
5X2X
 
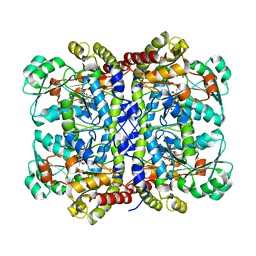 | |
7F8K
 
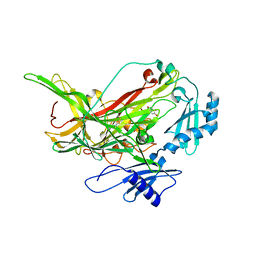 | |
8QMH
 
 | | Crystal structure of RNA G2C4 repeats in complex with small synthetic molecule ANP77 | | 分子名称: | 3-(7-azanyl-1,8-naphthyridin-2-yl)-2-[(7-azanyl-1,8-naphthyridin-2-yl)methyl]-~{N}-(3-azanylpropyl)propanamide, CHLORIDE ION, RNA (5'-R(*GP*GP*CP*CP*CP*C)-3') | | 著者 | Kiliszek, A, Ryczek, M, Blaszczyk, L. | | 登録日 | 2023-09-22 | | 公開日 | 2024-05-01 | | 最終更新日 | 2024-07-03 | | 実験手法 | X-RAY DIFFRACTION (1.1 Å) | | 主引用文献 | Antisense RNA C9orf72 hexanucleotide repeat associated with amyotrophic lateral sclerosis and frontotemporal dementia forms a triplex-like structure and binds small synthetic ligand.
Nucleic Acids Res., 52, 2024
|
|
8QMI
 
 | | Crystal structure of RNA G2C4 repeats - native model | | 分子名称: | MAGNESIUM ION, RNA (5'-R(*GP*GP*CP*CP*CP*C)-3') | | 著者 | Blaszczyk, L, Kiliszek, A. | | 登録日 | 2023-09-22 | | 公開日 | 2024-05-01 | | 最終更新日 | 2024-07-03 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Antisense RNA C9orf72 hexanucleotide repeat associated with amyotrophic lateral sclerosis and frontotemporal dementia forms a triplex-like structure and binds small synthetic ligand.
Nucleic Acids Res., 52, 2024
|
|
3RP2
 
 | | THE STRUCTURE OF RAT MAST CELL PROTEASE II AT 1.9-ANGSTROMS RESOLUTION | | 分子名称: | RAT MAST CELL PROTEASE II | | 著者 | Reynolds, R, Remington, S, Weaver, L, Fischer, R, Anderson, W, Ammon, H, Matthews, B. | | 登録日 | 1984-09-10 | | 公開日 | 1984-10-29 | | 最終更新日 | 2017-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | The structure of rat mast cell protease II at 1.9-A resolution.
Biochemistry, 27, 1988
|
|
6KZ7
 
 | | The crystal structure of BAF155 SWIRM domain and N-terminal elongated hSNF5 RPT1 domain complex: Chromatin remodeling complex | | 分子名称: | SWI/SNF complex subunit SMARCC1, SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily B member 1 | | 著者 | Lee, W, Han, J, Kim, I, Park, J.H, Joo, K, Lee, J, Suh, J.Y. | | 登録日 | 2019-09-23 | | 公開日 | 2020-07-08 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.28 Å) | | 主引用文献 | A Coil-to-Helix Transition Serves as a Binding Motif for hSNF5 and BAF155 Interaction.
Int J Mol Sci, 21, 2020
|
|
2SFP
 
 | |
1SFT
 
 | | ALANINE RACEMASE | | 分子名称: | ACETATE ION, ALANINE RACEMASE, PYRIDOXAL-5'-PHOSPHATE | | 著者 | Shaw, J.P, Petsko, G.A, Ringe, D. | | 登録日 | 1996-09-20 | | 公開日 | 1997-02-12 | | 最終更新日 | 2024-06-05 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Determination of the structure of alanine racemase from Bacillus stearothermophilus at 1.9-A resolution.
Biochemistry, 36, 1997
|
|
1KP4
 
 | |
