7E8D
 
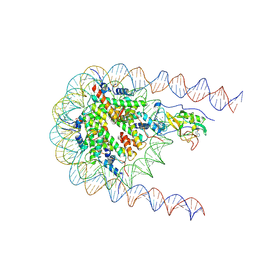 | | NSD2 E1099K mutant bound to nucleosome | | 分子名称: | DNA (185-MER), Histone H2A type 1, Histone H2B type 1-J, ... | | 著者 | Sengoku, T, Sato, K, Nishizawa, T, Nureki, O, Ogata, K. | | 登録日 | 2021-03-01 | | 公開日 | 2021-11-10 | | 最終更新日 | 2024-06-05 | | 実験手法 | ELECTRON MICROSCOPY (2.8 Å) | | 主引用文献 | Structural basis of the regulation of the normal and oncogenic methylation of nucleosomal histone H3 Lys36 by NSD2.
Nat Commun, 12, 2021
|
|
7BWO
 
 | |
7BWE
 
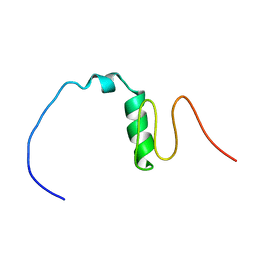 | |
5J5D
 
 | |
6TS8
 
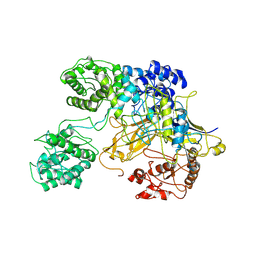 | | Chaetomium thermophilum UDP-Glucose Glucosyl Transferase (UGGT) double cysteine mutant G177C/A786C. | | 分子名称: | UDP-glucose-glycoprotein glucosyltransferase-like protein | | 著者 | Roversi, P, Zitzmann, N, Ibba, R, Hensen, M, Chandran, A. | | 登録日 | 2019-12-20 | | 公開日 | 2020-10-28 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (4.6 Å) | | 主引用文献 | Clamping, bending, and twisting inter-domain motions in the misfold-recognizing portion of UDP-glucose: Glycoprotein glucosyltransferase.
Structure, 29, 2021
|
|
1ZQT
 
 | |
3K5J
 
 | |
3NL9
 
 | |
3HSA
 
 | |
5GS8
 
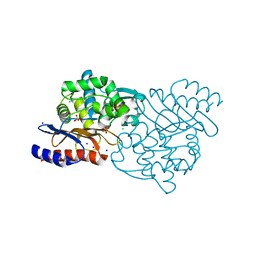 | | Crystal structure of TLA-3 extended-spectrum beta-lactamase | | 分子名称: | Beta-lactamase, CHLORIDE ION, SODIUM ION, ... | | 著者 | Wachino, J, Jin, W, Arakawa, Y. | | 登録日 | 2016-08-14 | | 公開日 | 2017-07-12 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.59 Å) | | 主引用文献 | Structural Insights into the TLA-3 Extended-Spectrum beta-Lactamase and Its Inhibition by Avibactam and OP0595.
Antimicrob. Agents Chemother., 61, 2017
|
|
5GWA
 
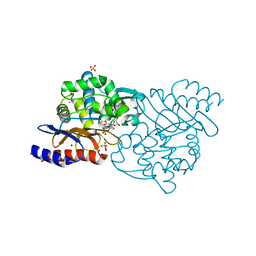 | | Crystal structure of TLA-3 extended-spectrum beta-lactamase in a complex with avibactam | | 分子名称: | (2S,5R)-1-formyl-5-[(sulfooxy)amino]piperidine-2-carboxamide, Beta-lactamase, CHLORIDE ION, ... | | 著者 | Wachino, J, Jin, W, Arakawa, Y. | | 登録日 | 2016-09-09 | | 公開日 | 2017-07-12 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (1.59 Å) | | 主引用文献 | Structural Insights into the TLA-3 Extended-Spectrum beta-Lactamase and Its Inhibition by Avibactam and OP0595.
Antimicrob. Agents Chemother., 61, 2017
|
|
3KK7
 
 | |
6TRT
 
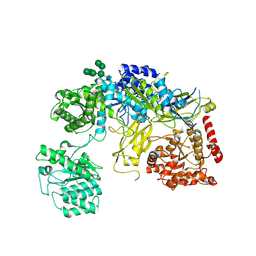 | | Chaetomium thermophilum UDP-Glucose Glucosyl Transferase (UGGT) double cysteine mutant S180C/T742C. | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, TERBIUM(III) ION, UDP-glucose-glycoprotein glucosyltransferase-like protein, ... | | 著者 | Roversi, P, Zitzmann, N, Ibba, R, Hensen, M. | | 登録日 | 2019-12-19 | | 公開日 | 2020-01-08 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (4.58 Å) | | 主引用文献 | Clamping, bending, and twisting inter-domain motions in the misfold-recognizing portion of UDP-glucose: Glycoprotein glucosyltransferase.
Structure, 29, 2021
|
|
6TRF
 
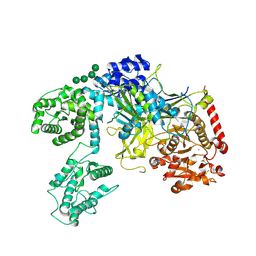 | | Chaetomium thermophilum UDP-Glucose Glucosyl Transferase (UGGT) purified from cells treated with kifunensine. | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, UDP-glucose-glycoprotein glucosyltransferase-like protein, ... | | 著者 | Roversi, P, Zitzmann, N. | | 登録日 | 2019-12-18 | | 公開日 | 2020-01-08 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (4.106 Å) | | 主引用文献 | Clamping, bending, and twisting inter-domain motions in the misfold-recognizing portion of UDP-glucose: Glycoprotein glucosyltransferase.
Structure, 29, 2021
|
|
6TS2
 
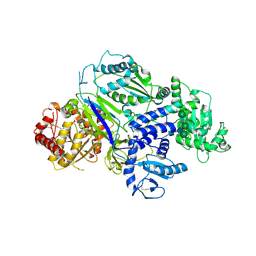 | | Truncated version of Chaetomium thermophilum UDP-Glucose Glucosyl Transferase (UGGT) lacking domain TRXL2 (417-650). | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, UDP-glucose-glycoprotein glucosyltransferase-like protein,UDP-glucose-glycoprotein glucosyltransferase-like protein, ... | | 著者 | Roversi, P, Zitzmann, N. | | 登録日 | 2019-12-19 | | 公開日 | 2020-01-08 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (5.74 Å) | | 主引用文献 | Clamping, bending, and twisting inter-domain motions in the misfold-recognizing portion of UDP-glucose: Glycoprotein glucosyltransferase.
Structure, 29, 2021
|
|
1BPZ
 
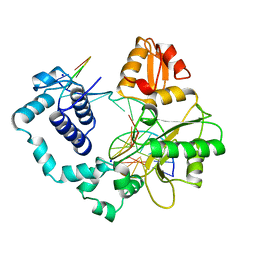 | | HUMAN DNA POLYMERASE BETA COMPLEXED WITH NICKED DNA | | 分子名称: | DNA (5'-D(*CP*CP*GP*AP*CP*CP*AP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3'), DNA (5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*TP*G)-3'), DNA (5'-D(*GP*TP*CP*GP*G)-3'), ... | | 著者 | Sawaya, M.R, Prasad, R, Wilson, S.H, Kraut, J, Pelletier, H. | | 登録日 | 1997-04-14 | | 公開日 | 1997-06-16 | | 最終更新日 | 2023-08-02 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Crystal structures of human DNA polymerase beta complexed with gapped and nicked DNA: evidence for an induced fit mechanism.
Biochemistry, 36, 1997
|
|
1BPY
 
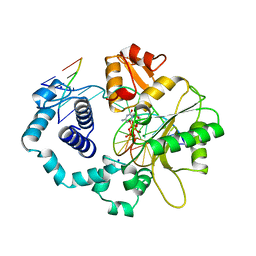 | | HUMAN DNA POLYMERASE BETA COMPLEXED WITH GAPPED DNA AND DDCTP | | 分子名称: | 2',3'-DIDEOXYCYTIDINE 5'-TRIPHOSPHATE, DNA (5'-D(*CP*CP*GP*AP*CP*GP*GP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3'), DNA (5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*DOC)-3'), ... | | 著者 | Sawaya, M.R, Pelletier, H, Prasad, R, Wilson, S.H, Kraut, J. | | 登録日 | 1997-04-15 | | 公開日 | 1997-06-16 | | 最終更新日 | 2023-08-02 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Crystal structures of human DNA polymerase beta complexed with gapped and nicked DNA: evidence for an induced fit mechanism.
Biochemistry, 36, 1997
|
|
1BPX
 
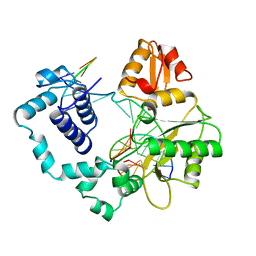 | | DNA POLYMERASE BETA/DNA COMPLEX | | 分子名称: | DNA (5'-D(*CP*CP*GP*AP*CP*GP*GP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3'), DNA (5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*C)-3'), DNA (5'-D(*GP*TP*CP*GP*G)-3'), ... | | 著者 | Sawaya, M.R, Prasad, R, Wilson, S.H, Kraut, J, Pelletier, H. | | 登録日 | 1997-04-11 | | 公開日 | 1997-06-16 | | 最終更新日 | 2023-08-02 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Crystal structures of human DNA polymerase beta complexed with gapped and nicked DNA: evidence for an induced fit mechanism.
Biochemistry, 36, 1997
|
|
6A8M
 
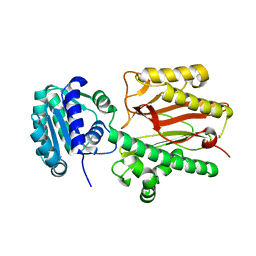 | | N-terminal domain of FACT complex subunit SPT16 from Eremothecium gossypii (Ashbya gossypii) | | 分子名称: | FACT complex subunit SPT16 | | 著者 | Gaur, N.K, Are, V.N, Durani, V, Ghosh, B, Kumar, A, Kulkarni, K, Makde, R.D. | | 登録日 | 2018-07-09 | | 公開日 | 2018-08-15 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Evolutionary conservation of protein dynamics: insights from all-atom molecular dynamics simulations of 'peptidase' domain of Spt16.
J.Biomol.Struct.Dyn., 2021
|
|
4D4Q
 
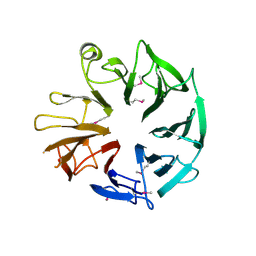 | | Crystal Structure of Kti13/AtS1 | | 分子名称: | PROTEIN ATS1 | | 著者 | Glatt, S, Mueller, C.W. | | 登録日 | 2014-10-30 | | 公開日 | 2015-01-14 | | 最終更新日 | 2017-03-29 | | 実験手法 | X-RAY DIFFRACTION (2.395 Å) | | 主引用文献 | Structure of the Kti11/Kti13 Heterodimer and its Double Role in Modifications of tRNA and Eukaryotic Elongation Factor 2.
Structure, 23, 2015
|
|
4D4O
 
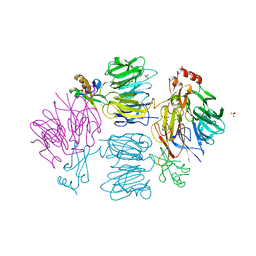 | |
4D4P
 
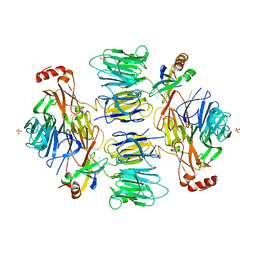 | |
4BQP
 
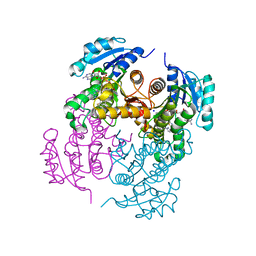 | | Mtb InhA complex with Methyl-thiazole compound 7 | | 分子名称: | (1S)-1-(5-{[1-(2,6-DIFLUOROBENZYL)-1H-PYRAZOL-3-YL]AMINO}-1,3,4-THIADIAZOL-2-YL)-1-(4-METHYL-1,3-THIAZOL-2-YL)ETHANOL, ENOYL-[ACYL-CARRIER-PROTEIN] REDUCTASE [NADH], NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | 著者 | Read, J.A, Gingell, H, Madhavapeddi, P, Shirude, P.S. | | 登録日 | 2013-05-31 | | 公開日 | 2013-12-11 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.89 Å) | | 主引用文献 | Methyl-Thiazoles: A Novel Mode of Inhibition with the Potential to Develop Novel Inhibitors Targeting Inha in Mycobacterium Tuberculosis.
J.Med.Chem., 56, 2013
|
|
6LEK
 
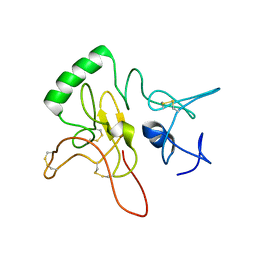 | | Tertiary structure of Barnacle cement protein MrCP20 | | 分子名称: | Cement protein-20k | | 著者 | Mohanram, H. | | 登録日 | 2019-11-25 | | 公開日 | 2020-01-15 | | 最終更新日 | 2024-10-16 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Three-dimensional structure of Megabalanus rosa Cement Protein 20 revealed by multi-dimensional NMR and molecular dynamics simulations.
Philos.Trans.R.Soc.Lond.B Biol.Sci., 374, 2019
|
|
3H0N
 
 | |
