5URY
 
 | | Crystal structure of Frizzled 5 CRD in complex with PAM | | 分子名称: | Frizzled-5, PALMITOLEIC ACID, alpha-L-fucopyranose-(1-3)-[alpha-L-fucopyranose-(1-4)][alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose | | 著者 | Mukund, S, Nile, A.H, Stanger, K, Hannoush, R.N, Wang, W. | | 登録日 | 2017-02-13 | | 公開日 | 2017-05-10 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (2.098 Å) | | 主引用文献 | Unsaturated fatty acyl recognition by Frizzled receptors mediates dimerization upon Wnt ligand binding.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
1DCH
 
 | | CRYSTAL STRUCTURE OF DCOH, A BIFUNCTIONAL, PROTEIN-BINDING TRANSCRIPTION COACTIVATOR | | 分子名称: | DCOH (DIMERIZATION COFACTOR OF HNF-1), SULFATE ION | | 著者 | Endrizzi, J.A, Cronk, J.D, Wang, W, Crabtree, G.R, Alber, T. | | 登録日 | 1995-01-24 | | 公開日 | 1996-03-08 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Crystal structure of DCoH, a bifunctional, protein-binding transcriptional coactivator.
Science, 268, 1995
|
|
5VRQ
 
 | | Crystal structure of Legionella pneumophila effector AnkC | | 分子名称: | Ankyrin repeat-containing protein | | 著者 | Kozlov, G, Wong, K, Wang, W, Skubak, P, Munoz-Escobar, J, Liu, Y, Pannu, N.S, Gehring, K, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | 登録日 | 2017-05-11 | | 公開日 | 2017-11-29 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (3.205 Å) | | 主引用文献 | Ankyrin repeats as a dimerization module.
Biochem. Biophys. Res. Commun., 495, 2018
|
|
5VZY
 
 | | Crystal structure of crenezumab Fab in complex with Abeta | | 分子名称: | Amyloid beta A4 protein, Crenezumab Fab heavy chain,Immunoglobulin gamma-1 heavy chain, Crenezumab Fab light chain,Immunoblobulin light chain | | 著者 | Ultsch, M, Wang, W. | | 登録日 | 2017-05-29 | | 公開日 | 2017-08-09 | | 最終更新日 | 2017-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.32 Å) | | 主引用文献 | Structure of Crenezumab Complex with Abeta Shows Loss of beta-Hairpin.
Sci Rep, 6, 2016
|
|
8IVM
 
 | | crystal structure of SulE mutant | | 分子名称: | Alpha/beta fold hydrolase, GLYCEROL, L(+)-TARTARIC ACID, ... | | 著者 | Liu, B, He, J, Ran, T, Wang, W. | | 登録日 | 2023-03-28 | | 公開日 | 2023-08-02 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (1.32 Å) | | 主引用文献 | Crystal structures of herbicide-detoxifying esterase reveal a lid loop affecting substrate binding and activity.
Nat Commun, 14, 2023
|
|
8IVS
 
 | | crystal structure of SulE mutant | | 分子名称: | Alpha/beta fold hydrolase, GLYCEROL, L(+)-TARTARIC ACID, ... | | 著者 | Liu, B, He, J, Ran, T, Wang, W. | | 登録日 | 2023-03-28 | | 公開日 | 2023-08-02 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (1.52 Å) | | 主引用文献 | Crystal structures of herbicide-detoxifying esterase reveal a lid loop affecting substrate binding and activity.
Nat Commun, 14, 2023
|
|
8J7J
 
 | | crystal structure of SulE mutant | | 分子名称: | Alpha/beta fold hydrolase, GLYCEROL, L(+)-TARTARIC ACID, ... | | 著者 | Liu, B, He, J, Ran, T, Wang, W. | | 登録日 | 2023-04-27 | | 公開日 | 2023-08-02 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (1.54 Å) | | 主引用文献 | Crystal structures of herbicide-detoxifying esterase reveal a lid loop affecting substrate binding and activity.
Nat Commun, 14, 2023
|
|
8IW6
 
 | | crystal structure of SulE mutant | | 分子名称: | Alpha/beta fold hydrolase, GLYCEROL, L(+)-TARTARIC ACID, ... | | 著者 | Liu, B, He, J, Ran, T, Wang, W. | | 登録日 | 2023-03-29 | | 公開日 | 2023-08-02 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (1.44 Å) | | 主引用文献 | Crystal structures of herbicide-detoxifying esterase reveal a lid loop affecting substrate binding and activity.
Nat Commun, 14, 2023
|
|
8IW3
 
 | | crystal structure of SulE mutant | | 分子名称: | 2-[[[[(4-CHLORO-6-METHOXY-2-PYRIMIDINYL)AMINO]CARBONYL]AMINO]SULFONYL]BENZOIC ACID ETHYL ESTER, Alpha/beta fold hydrolase, GLYCEROL, ... | | 著者 | Liu, B, He, J, Ran, T, Wang, W. | | 登録日 | 2023-03-29 | | 公開日 | 2023-08-02 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (1.56 Å) | | 主引用文献 | Crystal structures of herbicide-detoxifying esterase reveal a lid loop affecting substrate binding and activity.
Nat Commun, 14, 2023
|
|
8IVE
 
 | | crystal structure of SulE mutant | | 分子名称: | 2-[[[[(4-CHLORO-6-METHOXY-2-PYRIMIDINYL)AMINO]CARBONYL]AMINO]SULFONYL]BENZOIC ACID ETHYL ESTER, Alpha/beta fold hydrolase, GLYCEROL | | 著者 | Liu, B, He, J, Ran, T, Wang, W. | | 登録日 | 2023-03-27 | | 公開日 | 2023-08-02 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (1.44 Å) | | 主引用文献 | Crystal structures of herbicide-detoxifying esterase reveal a lid loop affecting substrate binding and activity.
Nat Commun, 14, 2023
|
|
8IVN
 
 | | crystal structure of SulE mutant | | 分子名称: | Alpha/beta fold hydrolase, GLYCEROL, L(+)-TARTARIC ACID, ... | | 著者 | Liu, B, He, J, Ran, T, Wang, W. | | 登録日 | 2023-03-28 | | 公開日 | 2023-08-02 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Crystal structures of herbicide-detoxifying esterase reveal a lid loop affecting substrate binding and activity.
Nat Commun, 14, 2023
|
|
8J7K
 
 | | crystal structure of SulE mutant | | 分子名称: | Alpha/beta fold hydrolase, GLYCEROL, L(+)-TARTARIC ACID, ... | | 著者 | Liu, B, He, J, Ran, T, Wang, W. | | 登録日 | 2023-04-27 | | 公開日 | 2023-08-02 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (1.36 Å) | | 主引用文献 | Crystal structures of herbicide-detoxifying esterase reveal a lid loop affecting substrate binding and activity.
Nat Commun, 14, 2023
|
|
8IVT
 
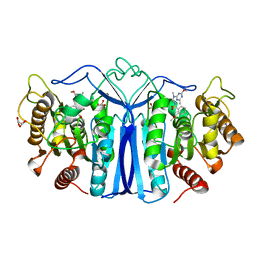 | | crystal structure of SulE mutant | | 分子名称: | Alpha/beta fold hydrolase, GLYCEROL, L(+)-TARTARIC ACID, ... | | 著者 | Liu, B, He, J, Ran, T, Wang, W. | | 登録日 | 2023-03-28 | | 公開日 | 2023-08-02 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (1.42 Å) | | 主引用文献 | Crystal structures of herbicide-detoxifying esterase reveal a lid loop affecting substrate binding and activity.
Nat Commun, 14, 2023
|
|
8J7G
 
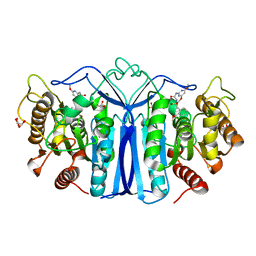 | | crystal structure of SulE mutant | | 分子名称: | Alpha/beta fold hydrolase, GLYCEROL, L(+)-TARTARIC ACID, ... | | 著者 | Liu, B, He, J, Ran, T, Wang, W. | | 登録日 | 2023-04-27 | | 公開日 | 2023-08-02 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (1.63 Å) | | 主引用文献 | Crystal structures of herbicide-detoxifying esterase reveal a lid loop affecting substrate binding and activity.
Nat Commun, 14, 2023
|
|
8GQR
 
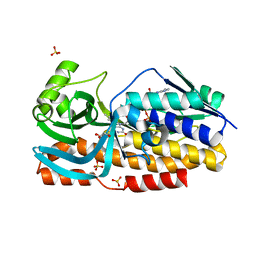 | | Crystal structure of VioD with FAD | | 分子名称: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, SULFATE ION, ... | | 著者 | Ran, T, Wang, W, Xu, M. | | 登録日 | 2022-08-30 | | 公開日 | 2023-03-29 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structural basis for substrate binding and catalytic mechanism of the key enzyme VioD in the violacein synthesis pathway.
Proteins, 91, 2023
|
|
8GOY
 
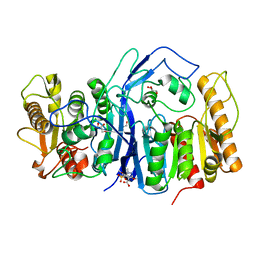 | | SulE P44R | | 分子名称: | 5-[(4,6-dimethoxypyrimidin-2-yl)carbamoylsulfamoyl]-1-methyl-pyrazole-4-carboxylic acid, Alpha/beta fold hydrolase, GLYCEROL | | 著者 | Liu, B, He, J, Ran, T, Wang, W. | | 登録日 | 2022-08-25 | | 公開日 | 2023-08-02 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (1.784 Å) | | 主引用文献 | Crystal structures of herbicide-detoxifying esterase reveal a lid loop affecting substrate binding and activity.
Nat Commun, 14, 2023
|
|
8GOL
 
 | | crystal structure of SulE | | 分子名称: | 2-[(4-chloranyl-6-methoxy-pyrimidin-2-yl)carbamoylsulfamoyl]benzoic acid, Alpha/beta fold hydrolase, GLYCEROL | | 著者 | Liu, B, Ran, T, Wang, W, He, J. | | 登録日 | 2022-08-25 | | 公開日 | 2023-08-02 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Crystal structures of herbicide-detoxifying esterase reveal a lid loop affecting substrate binding and activity.
Nat Commun, 14, 2023
|
|
8GP0
 
 | | crystal structure of SulE | | 分子名称: | Alpha/beta fold hydrolase, CITRIC ACID, GLYCEROL | | 著者 | Liu, B, Ran, T, wang, W, He, J. | | 登録日 | 2022-08-25 | | 公開日 | 2023-08-02 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (1.46 Å) | | 主引用文献 | Crystal structures of herbicide-detoxifying esterase reveal a lid loop affecting substrate binding and activity.
Nat Commun, 14, 2023
|
|
5YIM
 
 | |
8IVH
 
 | | crystal structure of SulE mutant | | 分子名称: | Alpha/beta fold hydrolase, GLYCEROL, L(+)-TARTARIC ACID, ... | | 著者 | Liu, B, He, J, Ran, T, Wang, W. | | 登録日 | 2023-03-27 | | 公開日 | 2024-04-10 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | crystal structure of SulE mutant
To Be Published
|
|
8IW8
 
 | |
8J7I
 
 | | crystal structure of SulE mutant | | 分子名称: | Alpha/beta fold hydrolase, GLYCEROL, L(+)-TARTARIC ACID, ... | | 著者 | Liu, B, He, J, Ran, T, Wang, W. | | 登録日 | 2023-04-27 | | 公開日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (1.54 Å) | | 主引用文献 | crystal structure of SulE mutant
To Be Published
|
|
8J7L
 
 | | crystal structure of SulE mutant | | 分子名称: | Alpha/beta fold hydrolase, GLYCEROL, L(+)-TARTARIC ACID, ... | | 著者 | Liu, B, He, J, Ran, T, Wang, W. | | 登録日 | 2023-04-27 | | 公開日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (1.59 Å) | | 主引用文献 | crystal structure of SulE mutant
To Be Published
|
|
6CQE
 
 | | Crystal structure of HPK1 kinase domain S171A mutant | | 分子名称: | Mitogen-activated protein kinase kinase kinase kinase 1 | | 著者 | Wu, P, Lehoux, I, Mortara, K, Franke, Y, Wang, W. | | 登録日 | 2018-03-15 | | 公開日 | 2018-12-19 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.886 Å) | | 主引用文献 | Hematopoietic Progenitor Kinase-1 Structure in a Domain-Swapped Dimer.
Structure, 27, 2019
|
|
8HEH
 
 | | Crystal structure of GCN5-related N-acetyltransferase 05790 | | 分子名称: | COENZYME A, GLYCEROL, GNAT family N-acetyltransferase | | 著者 | Xu, M.X, Ran, T.T, Wang, W. | | 登録日 | 2022-11-08 | | 公開日 | 2022-12-21 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Crystal structure of prodigiosin binding protein PgbP, a GNAT family protein, in Serratia marcescens FS14.
Biochem.Biophys.Res.Commun., 640, 2022
|
|
