5ELK
 
 | | Crystal structure of mouse Unkempt zinc fingers 4-6 (ZnF4-6), bound to RNA | | 分子名称: | RING finger protein unkempt homolog, RNA, ZINC ION | | 著者 | Teplova, M, Murn, J, Zarnack, K, Shi, Y, Patel, D.J. | | 登録日 | 2015-11-04 | | 公開日 | 2015-12-09 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Recognition of distinct RNA motifs by the clustered CCCH zinc fingers of neuronal protein Unkempt.
Nat.Struct.Mol.Biol., 23, 2016
|
|
5F1B
 
 | | Structural basis of Ebola virus entry: viral glycoprotein bound to its endosomal receptor Niemann-Pick C1 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GP1, GP2, ... | | 著者 | Wang, H, Shi, Y, Song, J, Qi, J, Lu, G, Yan, J, Gao, G.F. | | 登録日 | 2015-11-30 | | 公開日 | 2016-01-20 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Ebola Viral Glycoprotein Bound to Its Endosomal Receptor Niemann-Pick C1.
Cell, 164, 2016
|
|
5F18
 
 | | Structural basis of Ebola virus entry: viral glycoprotein bound to its endosomal receptor Niemann-Pick C1 | | 分子名称: | Niemann-Pick C1 protein | | 著者 | Wang, H, Shi, Y, Song, J, Qi, J, Lu, G, Yan, J, Gao, G.F. | | 登録日 | 2015-11-30 | | 公開日 | 2016-01-20 | | 最終更新日 | 2016-01-27 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Ebola Viral Glycoprotein Bound to Its Endosomal Receptor Niemann-Pick C1.
Cell, 164, 2016
|
|
5X4R
 
 | | Structure of the N-terminal domain (NTD) of MERS-CoV spike protein | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, S protein | | 著者 | Yuan, Y, Zhang, Y, Qi, J, Shi, Y, Gao, G.F. | | 登録日 | 2017-02-14 | | 公開日 | 2017-05-03 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Cryo-EM structures of MERS-CoV and SARS-CoV spike glycoproteins reveal the dynamic receptor binding domains
Nat Commun, 8, 2017
|
|
5X58
 
 | | Prefusion structure of SARS-CoV spike glycoprotein, conformation 1 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | 著者 | Yuan, Y, Cao, D, Zhang, Y, Ma, J, Qi, J, Wang, Q, Lu, G, Wu, Y, Yan, J, Shi, Y, Zhang, X, Gao, G.F. | | 登録日 | 2017-02-15 | | 公開日 | 2017-05-03 | | 最終更新日 | 2020-07-29 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Cryo-EM structures of MERS-CoV and SARS-CoV spike glycoproteins reveal the dynamic receptor binding domains
Nat Commun, 8, 2017
|
|
5X5B
 
 | | Prefusion structure of SARS-CoV spike glycoprotein, conformation 2 | | 分子名称: | Spike glycoprotein | | 著者 | Yuan, Y, Cao, D, Zhang, Y, Ma, J, Qi, J, Wang, Q, Lu, G, Wu, Y, Yan, J, Shi, Y, Zhang, X, Gao, G.F. | | 登録日 | 2017-02-15 | | 公開日 | 2017-05-03 | | 最終更新日 | 2017-05-24 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Cryo-EM structures of MERS-CoV and SARS-CoV spike glycoproteins reveal the dynamic receptor binding domains
Nat Commun, 8, 2017
|
|
5X59
 
 | | Prefusion structure of MERS-CoV spike glycoprotein, three-fold symmetry | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, S protein | | 著者 | Yuan, Y, Cao, D, Zhang, Y, Ma, J, Qi, J, Wang, Q, Lu, G, Wu, Y, Yan, J, Shi, Y, Zhang, X, Gao, G.F. | | 登録日 | 2017-02-15 | | 公開日 | 2017-05-03 | | 最終更新日 | 2020-07-29 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Cryo-EM structures of MERS-CoV and SARS-CoV spike glycoproteins reveal the dynamic receptor binding domains
Nat Commun, 8, 2017
|
|
5X5C
 
 | | Prefusion structure of MERS-CoV spike glycoprotein, conformation 1 | | 分子名称: | S protein | | 著者 | Yuan, Y, Cao, D, Zhang, Y, Ma, J, Qi, J, Wang, Q, Lu, G, Wu, Y, Yan, J, Shi, Y, Zhang, X, Gao, G.F. | | 登録日 | 2017-02-15 | | 公開日 | 2017-05-03 | | 最終更新日 | 2017-05-24 | | 実験手法 | ELECTRON MICROSCOPY (4.1 Å) | | 主引用文献 | Cryo-EM structures of MERS-CoV and SARS-CoV spike glycoproteins reveal the dynamic receptor binding domains
Nat Commun, 8, 2017
|
|
5X4S
 
 | | Structure of the N-terminal domain (NTD)of SARS-CoV spike protein | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | 著者 | Yuan, Y, Zhang, Y, Qi, J, Shi, Y, Gao, G.F. | | 登録日 | 2017-02-14 | | 公開日 | 2017-05-03 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Cryo-EM structures of MERS-CoV and SARS-CoV spike glycoproteins reveal the dynamic receptor binding domains
Nat Commun, 8, 2017
|
|
5X5F
 
 | | Prefusion structure of MERS-CoV spike glycoprotein, conformation 2 | | 分子名称: | S protein | | 著者 | Yuan, Y, Cao, D, Zhang, Y, Ma, J, Qi, J, Wang, Q, Lu, G, Wu, Y, Yan, J, Shi, Y, Zhang, X, Gao, G.F. | | 登録日 | 2017-02-15 | | 公開日 | 2017-05-03 | | 最終更新日 | 2017-05-24 | | 実験手法 | ELECTRON MICROSCOPY (4.2 Å) | | 主引用文献 | Cryo-EM structures of MERS-CoV and SARS-CoV spike glycoproteins reveal the dynamic receptor binding domains
Nat Commun, 8, 2017
|
|
5GM6
 
 | | Cryo-EM structure of the activated spliceosome (Bact complex) at 3.5 angstrom resolution | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Cold sensitive U2 snRNA suppressor 1, GUANOSINE-5'-TRIPHOSPHATE, ... | | 著者 | Yan, C, Wan, R, Bai, R, Huang, G, Shi, Y. | | 登録日 | 2016-07-12 | | 公開日 | 2016-09-21 | | 最終更新日 | 2019-11-06 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | Structure of a yeast activated spliceosome at 3.5 angstrom resolution
Science, 353, 2016
|
|
5GMK
 
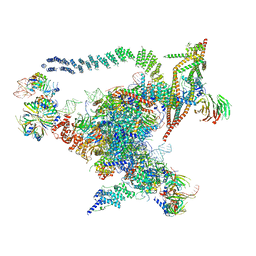 | | Cryo-EM structure of the Catalytic Step I spliceosome (C complex) at 3.4 angstrom resolution | | 分子名称: | 5'-Exon, 5'-Splicing Site, GUANOSINE-5'-TRIPHOSPHATE, ... | | 著者 | Wan, R, Yan, C, Bai, R, Huang, G, Shi, Y. | | 登録日 | 2016-07-14 | | 公開日 | 2016-08-17 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Structure of a yeast catalytic step I spliceosome at 3.4 angstrom resolution
Science, 353, 2016
|
|
7VPX
 
 | |
7WB4
 
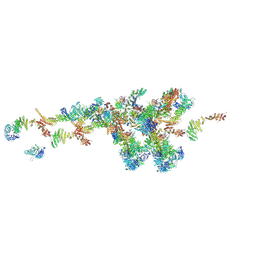 | | Cryo-EM structure of the NR subunit from X. laevis NPC | | 分子名称: | GATOR complex protein SEC13, MGC154553 protein, MGC83295 protein, ... | | 著者 | Huang, G, Zhan, X, Shi, Y. | | 登録日 | 2021-12-15 | | 公開日 | 2022-03-02 | | 最終更新日 | 2022-04-20 | | 実験手法 | ELECTRON MICROSCOPY (5.6 Å) | | 主引用文献 | Cryo-EM structure of the nuclear ring from Xenopus laevis nuclear pore complex.
Cell Res., 32, 2022
|
|
7WKK
 
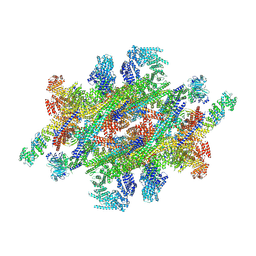 | | Cryo-EM structure of the IR subunit from X. laevis NPC | | 分子名称: | Aaas-prov protein, IL4I1 protein, MGC83295 protein, ... | | 著者 | Huang, G, Zhan, X, Shi, Y. | | 登録日 | 2022-01-10 | | 公開日 | 2022-03-30 | | 最終更新日 | 2022-05-18 | | 実験手法 | ELECTRON MICROSCOPY (4.2 Å) | | 主引用文献 | Cryo-EM structure of the inner ring from the Xenopus laevis nuclear pore complex.
Cell Res., 32, 2022
|
|
7WOL
 
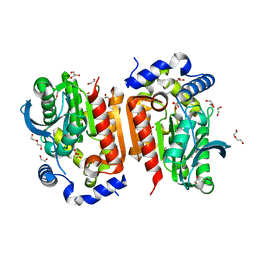 | | Crystal structure of lipase TrLipB from Thermomocrobium roseum | | 分子名称: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | 著者 | Zhang, L, Fang, Y, Shi, Y, Gu, Z, Xin, Y. | | 登録日 | 2022-01-21 | | 公開日 | 2023-01-25 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystal structure of lipase TrLipB from Thermomocrobium roseum
To Be Published
|
|
5VRT
 
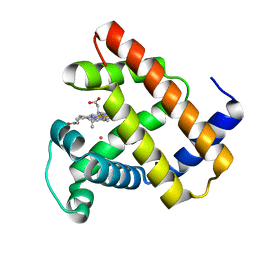 | | Nonheme Iron Replacement in a Biosynthetic Nitric Oxide Reductase Model Performing O2 Reduction to Water: Co-bound FeBMb | | 分子名称: | COBALT (II) ION, Myoglobin, PROTOPORPHYRIN IX CONTAINING FE | | 著者 | Reed, J, Shi, Y, Zhu, Q, Chakraborty, S, Mirs, E.N, Petrik, I.D, Bhagi-Damodaran, A, Ross, M, Moenne-Loccoz, P, Zhang, Y, Lu, Y. | | 登録日 | 2017-05-11 | | 公開日 | 2017-08-16 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.995 Å) | | 主引用文献 | Manganese and Cobalt in the Nonheme-Metal-Binding Site of a Biosynthetic Model of Heme-Copper Oxidase Superfamily Confer Oxidase Activity through Redox-Inactive Mechanism.
J. Am. Chem. Soc., 139, 2017
|
|
6KUK
 
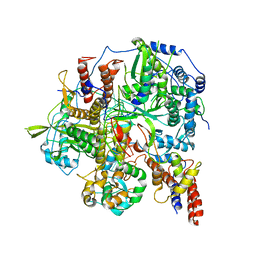 | | Structure of influenza D virus polymerase bound to vRNA promoter in mode A conformation (class A1) | | 分子名称: | 3'-vRNA, 5'-vRNA, Polymerase 3, ... | | 著者 | Peng, Q, Peng, R, Qi, J, Gao, G.F, Shi, Y. | | 登録日 | 2019-09-02 | | 公開日 | 2019-10-02 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | Structural insight into RNA synthesis by influenza D polymerase.
Nat Microbiol, 4, 2019
|
|
6KUR
 
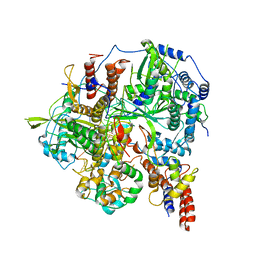 | | Structure of influenza D virus polymerase bound to vRNA promoter in Mode B conformation (Class B1) | | 分子名称: | 3'-vRNA, 5'-vRNA, Polymerase 3, ... | | 著者 | Peng, Q, Peng, R, Qi, J, Gao, G.F, Shi, Y. | | 登録日 | 2019-09-02 | | 公開日 | 2019-10-02 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Structural insight into RNA synthesis by influenza D polymerase.
Nat Microbiol, 4, 2019
|
|
6KUP
 
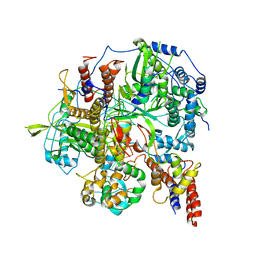 | | Structure of influenza D virus polymerase bound to vRNA promoter in Mode A conformation(Class A2) | | 分子名称: | 3'-vRNA, 5'-vRNA, Polymerase 3, ... | | 著者 | Peng, Q, Peng, R, Qi, J, Gao, G.F, Shi, Y. | | 登録日 | 2019-09-02 | | 公開日 | 2019-10-02 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (4.3 Å) | | 主引用文献 | Structural insight into RNA synthesis by influenza D polymerase.
Nat Microbiol, 4, 2019
|
|
6KUV
 
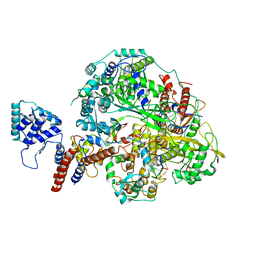 | | Structure of influenza D virus polymerase bound to cRNA promoter in class 2 | | 分子名称: | 3'-cRNA, 5'-cRNA, Polymerase 3, ... | | 著者 | Peng, Q, Peng, R, Qi, J, Gao, G.F, Shi, Y. | | 登録日 | 2019-09-02 | | 公開日 | 2019-10-02 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (4.1 Å) | | 主引用文献 | Structural insight into RNA synthesis by influenza D polymerase.
Nat Microbiol, 4, 2019
|
|
6KUT
 
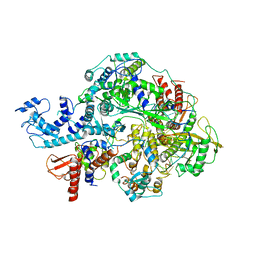 | | Structure of influenza D virus polymerase bound to vRNA promoter in Mode B conformation (Class B2) | | 分子名称: | 3'-vRNA, 5'-vRNA, Polymerase 3, ... | | 著者 | Peng, Q, Peng, R, Qi, J, Gao, G.F, Shi, Y. | | 登録日 | 2019-09-02 | | 公開日 | 2019-10-02 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (4.1 Å) | | 主引用文献 | Structural insight into RNA synthesis by influenza D polymerase.
Nat Microbiol, 4, 2019
|
|
6KV5
 
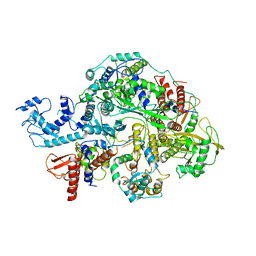 | | Structure of influenza D virus apo polymerase | | 分子名称: | Polymerase 3, Polymerase PB2, RNA-directed RNA polymerase catalytic subunit | | 著者 | Peng, Q, Peng, R, Qi, J, Gao, G.F, Shi, Y. | | 登録日 | 2019-09-03 | | 公開日 | 2019-10-02 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (4.6 Å) | | 主引用文献 | Structural insight into RNA synthesis by influenza D polymerase.
Nat Microbiol, 4, 2019
|
|
5XLP
 
 | |
5XGR
 
 | | Structure of the S1 subunit C-terminal domain from bat-derived coronavirus HKU5 spike protein | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1 | | 著者 | Xue, H, Qi, J, Song, H, Qihui, W, Shi, Y, Gao, G.F. | | 登録日 | 2017-04-16 | | 公開日 | 2017-05-10 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structure of the S1 subunit C-terminal domain from bat-derived coronavirus HKU5 spike protein
Virology, 507, 2017
|
|
