8VBK
 
 | | Structure of bovine anti-HIV Fab ElsE5 | | 分子名称: | Bovine Fab ElsE5 heavy chain, Bovine Fab ElsE5 light chain | | 著者 | Stanfield, R.L, Wilson, I.A. | | 登録日 | 2023-12-12 | | 公開日 | 2024-09-04 | | 最終更新日 | 2024-11-13 | | 実験手法 | X-RAY DIFFRACTION (1.89 Å) | | 主引用文献 | Immunization of cows with HIV envelope trimers generates broadly neutralizing antibodies to the V2-apex from the ultralong CDRH3 repertoire.
Plos Pathog., 20, 2024
|
|
8VBJ
 
 | | Structure of bovine anti-HIV Fab ElsE2 | | 分子名称: | Bovine Fab ElsE2 heavy chain, Bovine Fab ElsE2 light chain, POTASSIUM ION, ... | | 著者 | Stanfield, R.L, Wilson, I.A. | | 登録日 | 2023-12-12 | | 公開日 | 2024-09-04 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Immunization of cows with HIV envelope trimers generates broadly neutralizing antibodies to the V2-apex from the ultralong CDRH3 repertoire.
Plos Pathog., 20, 2024
|
|
8VBP
 
 | | Structure of bovine anti-HIV Fab Bess4 | | 分子名称: | Bovine Fab Bess4 heavy chain, Bovine Fab Bess4 light chain | | 著者 | Stanfield, R.L, Wilson, I.A. | | 登録日 | 2023-12-12 | | 公開日 | 2024-09-04 | | 最終更新日 | 2024-11-20 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Immunization of cows with HIV envelope trimers generates broadly neutralizing antibodies to the V2-apex from the ultralong CDRH3 repertoire.
Plos Pathog., 20, 2024
|
|
8VBN
 
 | | Structure of bovine anti-HIV Fab ElsE8 | | 分子名称: | Bovine Fab ElsE8 heavy chain, Bovine Fab ElsE8 light chain | | 著者 | Stanfield, R.L, Wilson, I.A. | | 登録日 | 2023-12-12 | | 公開日 | 2024-09-04 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (1.83 Å) | | 主引用文献 | Immunization of cows with HIV envelope trimers generates broadly neutralizing antibodies to the V2-apex from the ultralong CDRH3 repertoire.
Plos Pathog., 20, 2024
|
|
8V4I
 
 | | Structure of bovine anti-HIV Fab ElsE1 | | 分子名称: | ElsE1 Fab heavy chain, ElsE1 Fab light chain | | 著者 | Stanfield, R.L, Wilson, I.A. | | 登録日 | 2023-11-29 | | 公開日 | 2024-09-04 | | 最終更新日 | 2025-03-05 | | 実験手法 | X-RAY DIFFRACTION (1.81 Å) | | 主引用文献 | Immunization of cows with HIV envelope trimers generates broadly neutralizing antibodies to the V2-apex from the ultralong CDRH3 repertoire.
Plos Pathog., 20, 2024
|
|
8VBO
 
 | | Structure of bovine anti-HIV Fab ElsE9 | | 分子名称: | Bovine Fab ElsE9 heavy chain, Bovine Fab ElsE9 light chain, GLYCEROL, ... | | 著者 | Stanfield, R.L, Wilson, I.A. | | 登録日 | 2023-12-12 | | 公開日 | 2024-09-04 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Immunization of cows with HIV envelope trimers generates broadly neutralizing antibodies to the V2-apex from the ultralong CDRH3 repertoire.
Plos Pathog., 20, 2024
|
|
8VBQ
 
 | | Structure of bovine anti-HIV Fab Bess7 | | 分子名称: | Bovine Fab Bess7 heavy chain, Bovine Fab Bess7 light chain | | 著者 | Stanfield, R.L, Wilson, I.A. | | 登録日 | 2023-12-12 | | 公開日 | 2024-09-04 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Immunization of cows with HIV envelope trimers generates broadly neutralizing antibodies to the V2-apex from the ultralong CDRH3 repertoire.
Plos Pathog., 20, 2024
|
|
4N4A
 
 | | Cystal structure of Cap-specific mRNA (nucleoside-2'-O-)-methyltransferase 1 | | 分子名称: | Cap-specific mRNA (nucleoside-2'-O-)-methyltransferase 1 | | 著者 | Smietanski, M, Werener, M, Purta, E, Kaminska, K.H, Stepinski, J, Darzynkiewicz, E, Nowotny, M, Bujnicki, J.M. | | 登録日 | 2013-10-08 | | 公開日 | 2014-01-22 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | Structural analysis of human 2'-O-ribose methyltransferases involved in mRNA cap structure formation.
Nat Commun, 5, 2014
|
|
8VI1
 
 | | Crystal structure of c-Met-D1228N in complex with KIN-7615 | | 分子名称: | Hepatocyte growth factor receptor, N-(3,5-difluoro-4-{[6-(2-hydroxyethoxy)-7-methoxyquinolin-4-yl]oxy}phenyl)-4-methoxypyridine-3-carboxamide | | 著者 | Ouyang, X, Kania, R, Cox, J. | | 登録日 | 2024-01-02 | | 公開日 | 2025-04-02 | | 最終更新日 | 2025-06-25 | | 実験手法 | X-RAY DIFFRACTION (3.11 Å) | | 主引用文献 | Discovery of KIN-8741, a Highly Selective Type IIb c-Met Kinase Inhibitor with Broad Mutation Coverage and Quality Drug-Like Properties for the Treatment of Cancer.
J.Med.Chem., 68, 2025
|
|
6ORE
 
 | | Release complex 70S | | 分子名称: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | 著者 | Fu, Z. | | 登録日 | 2019-04-30 | | 公開日 | 2019-06-19 | | 最終更新日 | 2025-03-19 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | The structural basis for release-factor activation during translation termination revealed by time-resolved cryogenic electron microscopy.
Nat Commun, 10, 2019
|
|
3EP2
 
 | | Model of Phe-tRNA(Phe) in the ribosomal pre-accommodated state revealed by cryo-EM | | 分子名称: | 30S ribosomal protein S12, 50S ribosomal protein L11, Elongation factor Tu, ... | | 著者 | Frank, J, Li, W, Agirrezabala, X. | | 登録日 | 2008-09-29 | | 公開日 | 2008-12-16 | | 最終更新日 | 2024-02-21 | | 実験手法 | ELECTRON MICROSCOPY (9 Å) | | 主引用文献 | Recognition of aminoacyl-tRNA: a common molecular mechanism revealed by cryo-EM.
Embo J., 27, 2008
|
|
5LY9
 
 | | Structure of MITat 1.1 | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLYCEROL, Variant surface glycoprotein MITAT 1.1, ... | | 著者 | Schaefer, C, Bartossek, T, Jones, N, Kuper, J, Kisker, C, Engstler, M. | | 登録日 | 2016-09-26 | | 公開日 | 2017-09-20 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Structural basis for the shielding function of the dynamic trypanosome variant surface glycoprotein coat.
Nat Microbiol, 2, 2017
|
|
7AIT
 
 | | Crystal structure of Torpedo Californica acetylcholinesterase in complex with 7-[(3-Chloro-6,7,10,11-tetrahydro-9-methyl-7,11-methanocycloocta[b]quinolin-12-yl)amino]-N-(4-hydroxy-3-methoxybenzyl)heptanamide | | 分子名称: | 7-[(3-Chloro-6,7,10,11-tetrahydro-9-methyl-7,11-methanocycloocta[b]quinolin-12-yl)amino]-N-(4-hydroxy-3-methoxybenzyl)heptanamide, Acetylcholinesterase | | 著者 | Coquelle, N, Colletier, J.P. | | 登録日 | 2020-09-28 | | 公開日 | 2021-10-06 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Discovery of a Potent Dual Inhibitor of Acetylcholinesterase and Butyrylcholinesterase with Antioxidant Activity that Alleviates Alzheimer-like Pathology in Old APP/PS1 Mice.
J.Med.Chem., 64, 2021
|
|
7AIS
 
 | | Crystal structure of Torpedo Californica acetylcholinesterase in complex with 6-[(3-Chloro-6,7,10,11-tetrahydro-9-methyl-7,11-methanocycloocta[b]quinolin-12-yl)amino]-N-(4-hydroxy-3-methoxybenzyl)hexanamide | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 6-[(3-Chloro-6,7,10,11-tetrahydro-9-methyl-7,11-methanocycloocta[b]quinolin-12-yl)amino]-N-(4-hydroxy-3-methoxybenzyl)hexanamide, Acetylcholinesterase, ... | | 著者 | Coquelle, N, Colletier, J.P. | | 登録日 | 2020-09-28 | | 公開日 | 2021-10-06 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Discovery of a Potent Dual Inhibitor of Acetylcholinesterase and Butyrylcholinesterase with Antioxidant Activity that Alleviates Alzheimer-like Pathology in Old APP/PS1 Mice.
J.Med.Chem., 64, 2021
|
|
7AIY
 
 | | Crystal structure of human butyrylcholinesterase in complex with 2-{1-[4-(12-Amino-3-chloro-6,7,10,11-tetrahydro-7,11-methanocycloocta[b]quinolin-9-yl)butyl]-1H-1,2,3-triazol-4-yl}-N-[4-hydroxy-3-methoxybenzyl]acetamide | | 分子名称: | 2-{1-[4-(12-Amino-3-chloro-6,7,10,11-tetrahydro-7,11-methanocycloocta[b]quinolin-9-yl)butyl]-1H-1,2,3-triazol-4-yl}-N-[4-hydroxy-3-methoxybenzyl]acetamide, Cholinesterase | | 著者 | Coquelle, N, Colletier, J.P. | | 登録日 | 2020-09-28 | | 公開日 | 2021-10-06 | | 最終更新日 | 2024-11-13 | | 実験手法 | X-RAY DIFFRACTION (2.937 Å) | | 主引用文献 | Discovery of a Potent Dual Inhibitor of Acetylcholinesterase and Butyrylcholinesterase with Antioxidant Activity that Alleviates Alzheimer-like Pathology in Old APP/PS1 Mice.
J.Med.Chem., 64, 2021
|
|
7AIU
 
 | | Crystal structure of Torpedo Californica acetylcholinesterase in complex with 8-[(3-Chloro-6,7,10,11-tetrahydro-9-methyl-7,11-methanocycloocta[b]quinolin-12-yl)amino]-N-(4-hydroxy-3-methoxybenzyl)octanamide | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 8-[(3-Chloro-6,7,10,11-tetrahydro-9-methyl-7,11-methanocycloocta[b]quinolin-12-yl)amino]-N-(4-hydroxy-3-methoxybenzyl)octanamide, Acetylcholinesterase, ... | | 著者 | Coquelle, N, Colletier, J.P. | | 登録日 | 2020-09-28 | | 公開日 | 2021-10-06 | | 最終更新日 | 2024-11-20 | | 実験手法 | X-RAY DIFFRACTION (1.996 Å) | | 主引用文献 | Discovery of a Potent Dual Inhibitor of Acetylcholinesterase and Butyrylcholinesterase with Antioxidant Activity that Alleviates Alzheimer-like Pathology in Old APP/PS1 Mice.
J.Med.Chem., 64, 2021
|
|
7AIX
 
 | | Crystal structure of Torpedo Californica acetylcholinesterase in complex with 2-{1-[4-(12-Amino-3-chloro-6,7,10,11-tetrahydro-7,11-methanocycloocta[b]quinolin-9-yl)butyl]-1H-1,2,3-triazol-4-yl}-N-[4-hydroxy-3-methoxybenzyl]acetamide | | 分子名称: | 2-{1-[4-(12-Amino-3-chloro-6,7,10,11-tetrahydro-7,11-methanocycloocta[b]quinolin-9-yl)butyl]-1H-1,2,3-triazol-4-yl}-N-[4-hydroxy-3-methoxybenzyl]acetamide, Acetylcholinesterase, CHLORIDE ION, ... | | 著者 | Coquelle, N, Colletier, J.P. | | 登録日 | 2020-09-28 | | 公開日 | 2021-10-06 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (1.86 Å) | | 主引用文献 | Discovery of a Potent Dual Inhibitor of Acetylcholinesterase and Butyrylcholinesterase with Antioxidant Activity that Alleviates Alzheimer-like Pathology in Old APP/PS1 Mice.
J.Med.Chem., 64, 2021
|
|
7AIW
 
 | | Crystal structure of Torpedo Californica acetylcholinesterase in complex with (E)-10-[(3-Chloro-6,7,10,11-tetrahydro-9-methyl-7,11-methanocycloocta[b]quinolin-12-yl)amino]-N-(4-hydroxy-3-methoxybenzyl)-6-decenamide | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, Acetylcholinesterase, ... | | 著者 | Coquelle, N, Colletier, J.P. | | 登録日 | 2020-09-28 | | 公開日 | 2021-10-06 | | 最終更新日 | 2024-11-20 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Discovery of a Potent Dual Inhibitor of Acetylcholinesterase and Butyrylcholinesterase with Antioxidant Activity that Alleviates Alzheimer-like Pathology in Old APP/PS1 Mice.
J.Med.Chem., 64, 2021
|
|
7AIV
 
 | | Crystal structure of Torpedo Californica acetylcholinesterase in complex with 4-{[(3-Chloro-6,7,10,11-tetrahydro-9-methyl-7,11-methanocycloocta[b]quinolin-12-yl)amino]methyl}-N-(4-hydroxy-3-methoxybenzyl)benzamide | | 分子名称: | 4-{[(3-Chloro-6,7,10,11-tetrahydro-9-methyl-7,11-methanocycloocta[b]quinolin-12-yl)amino]methyl}-N-(4-hydroxy-3-methoxybenzyl)benzamide, Acetylcholinesterase | | 著者 | Coquelle, N, Colletier, J.P. | | 登録日 | 2020-09-28 | | 公開日 | 2021-10-06 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (2.55 Å) | | 主引用文献 | Discovery of a Potent Dual Inhibitor of Acetylcholinesterase and Butyrylcholinesterase with Antioxidant Activity that Alleviates Alzheimer-like Pathology in Old APP/PS1 Mice.
J.Med.Chem., 64, 2021
|
|
6XLI
 
 | | CRYSTAL STRUCTURE OF ANTI-TAU ANTIBODY PT3 Fab+pT212/pT217-TAU PEPTIDE | | 分子名称: | GLYCEROL, PT3 Fab Heavy Chain, PT3 Fab Light Chain, ... | | 著者 | Malia, T.J, Teplyakov, A, Luo, J. | | 登録日 | 2020-06-28 | | 公開日 | 2020-09-23 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Discovery and Functional Characterization of hPT3, a Humanized Anti-Phospho Tau Selective Monoclonal Antibody.
J Alzheimers Dis, 77, 2020
|
|
5M4T
 
 | |
7SJS
 
 | |
4N49
 
 | | Cap-specific mRNA (nucleoside-2'-O-)-methyltransferase 1 Protein in complex with m7GpppG and SAM | | 分子名称: | 7N-METHYL-8-HYDROGUANOSINE-5'-TRIPHOSPHATE, Cap-specific mRNA (nucleoside-2'-O-)-methyltransferase 1, S-ADENOSYLMETHIONINE, ... | | 著者 | Smietanski, M, Werener, M, Purta, E, Kaminska, K.H, Stepinski, J, Darzynkiewicz, E, Nowotny, M, Bujnicki, J.M. | | 登録日 | 2013-10-08 | | 公開日 | 2014-01-22 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structural analysis of human 2'-O-ribose methyltransferases involved in mRNA cap structure formation.
Nat Commun, 5, 2014
|
|
6EHT
 
 | | Modulation of PCNA sliding surface by p15PAF suggests a suppressive mechanism for cisplatin-induced DNA lesion bypass by pol eta holoenzyme | | 分子名称: | DNA (5'-D(P*AP*TP*AP*CP*GP*AP*TP*GP*GP*G)-3'), DNA (5'-D(P*CP*CP*CP*AP*TP*CP*GP*TP*AP*T)-3'), PCNA-associated factor, ... | | 著者 | De March, M, Barrera-Vilarmau, S, Mentegari, E, Merino, N, Bressan, E, Maga, G, Crehuet, R, Onesti, S, Blanco, F.J, De Biasio, A. | | 登録日 | 2017-09-15 | | 公開日 | 2018-08-08 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | p15PAF binding to PCNA modulates the DNA sliding surface.
Nucleic Acids Res., 46, 2018
|
|
5K0M
 
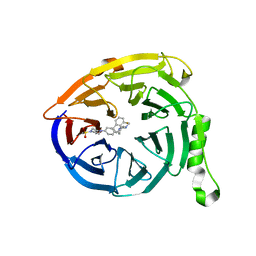 | | Targeting the PRC2 complex through a novel protein-protein interaction inhibitor of EED | | 分子名称: | (3R,4S)-1-[(1S)-7-fluoro-2,3-dihydro-1H-inden-1-yl]-N,N-dimethyl-4-{4-[4-(methylsulfonyl)piperazin-1-yl]phenyl}pyrrolidin-3-amine, Polycomb protein EED | | 著者 | Jakob, C.G, Bigelow, L.J, Zhu, H, Sun, C. | | 登録日 | 2016-05-17 | | 公開日 | 2017-01-25 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.83 Å) | | 主引用文献 | The EED protein-protein interaction inhibitor A-395 inactivates the PRC2 complex.
Nat. Chem. Biol., 13, 2017
|
|
