7KO6
 
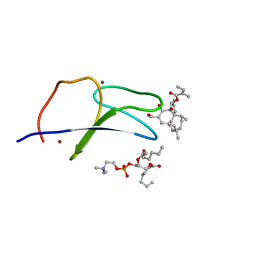 | | C1B domain of Protein kinase C in complex with ingenol-3-angelate and phosphocholine | | 分子名称: | (4S,7R)-7-(heptanoyloxy)-4-hydroxy-N,N,N-trimethyl-10-oxo-3,5,9-trioxa-4-phosphahexadecan-1-aminium 4-oxide, Protein kinase C delta type, ZINC ION, ... | | 著者 | Katti, S.S, Krieger, I.V. | | 登録日 | 2020-11-06 | | 公開日 | 2022-05-04 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structural anatomy of Protein Kinase C C1 domain interactions with diacylglycerol and other agonists.
Nat Commun, 13, 2022
|
|
7R40
 
 | |
3BM2
 
 | |
3BM1
 
 | |
4EO6
 
 | |
4EO8
 
 | | HCV NS5B polymerase inhibitors: Tri-substituted acylhydrazines as tertiary amide bioisosteres | | 分子名称: | 5-(3,3-dimethylbut-1-yn-1-yl)-3-{2,2-dimethyl-1-[(trans-4-methylcyclohexyl)carbonyl]hydrazinyl}thiophene-2-carboxylic acid, RNA-directed RNA polymerase | | 著者 | Appleby, T.C, Canales, E, Watkins, W.J. | | 登録日 | 2012-04-13 | | 公開日 | 2012-06-27 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.798 Å) | | 主引用文献 | Tri-substituted acylhydrazines as tertiary amide bioisosteres: HCV NS5B polymerase inhibitors.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
5K5O
 
 | | Structure of AspA-26mer DNA complex | | 分子名称: | AspA, DNA (26-MER) | | 著者 | Schumacher, M. | | 登録日 | 2016-05-23 | | 公開日 | 2016-06-15 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | Structures of archaeal DNA segregation machinery reveal bacterial and eukaryotic linkages.
Science, 349, 2015
|
|
5K5D
 
 | |
5K5R
 
 | | AspA-32mer DNA,crystal form 2 | | 分子名称: | AspA, DNA (32-MER), PHOSPHATE ION | | 著者 | Schumacher, M. | | 登録日 | 2016-05-23 | | 公開日 | 2016-06-15 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (3.09 Å) | | 主引用文献 | Structures of archaeal DNA segregation machinery reveal bacterial and eukaryotic linkages.
Science, 349, 2015
|
|
5I41
 
 | | Structure of the apo RacA DNA binding domain | | 分子名称: | Chromosome-anchoring protein RacA | | 著者 | schumacher, M.A. | | 登録日 | 2016-02-11 | | 公開日 | 2016-05-04 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Molecular insights into DNA binding and anchoring by the Bacillus subtilis sporulation kinetochore-like RacA protein.
Nucleic Acids Res., 44, 2016
|
|
1MRF
 
 | | PREPARATION, CHARACTERIZATION AND CRYSTALLIZATION OF AN ANTIBODY FAB FRAGMENT THAT RECOGNIZES RNA. CRYSTAL STRUCTURES OF NATIVE FAB AND THREE FAB-MONONUCLEOTIDE COMPLEXES | | 分子名称: | 2'-DEOXYINOSINE-5'-MONOPHOSPHATE, IGG2B-KAPPA JEL103 FAB (HEAVY CHAIN), IGG2B-KAPPA JEL103 FAB (LIGHT CHAIN), ... | | 著者 | Pokkuluri, P.R, Cygler, M. | | 登録日 | 1994-06-13 | | 公開日 | 1995-02-14 | | 最終更新日 | 2024-06-05 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Preparation, characterization and crystallization of an antibody Fab fragment that recognizes RNA. Crystal structures of native Fab and three Fab-mononucleotide complexes.
J.Mol.Biol., 243, 1994
|
|
4D8H
 
 | | Crystal structure of Symfoil-4P/PV2: de novo designed beta-trefoil architecture with symmetric primary structure, primitive version 2 (6xLeu / PV1) | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, SULFATE ION, de novo protein | | 著者 | Blaber, M, Longo, L. | | 登録日 | 2012-01-10 | | 公開日 | 2013-01-16 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.901 Å) | | 主引用文献 | Simplified protein design biased for prebiotic amino acids yields a foldable, halophilic protein.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
5IWS
 
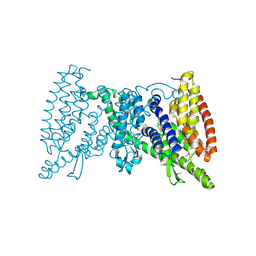 | | Crystal structure of the transporter MalT, the EIIC domain from the maltose-specific phosphotransferase system | | 分子名称: | Protein-N(Pi)-phosphohistidine-sugar phosphotransferase (Enzyme II of the phosphotransferase system) (PTS system glucose-specific IIBC component), alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | 著者 | McCoy, J.G, Ren, Z, Levin, E.J, Zhou, M, New York Consortium on Membrane Protein Structure (NYCOMPS) | | 登録日 | 2016-03-22 | | 公開日 | 2016-05-25 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (2.551 Å) | | 主引用文献 | The Structure of a Sugar Transporter of the Glucose EIIC Superfamily Provides Insight into the Elevator Mechanism of Membrane Transport.
Structure, 24, 2016
|
|
5K5Q
 
 | |
5K5Z
 
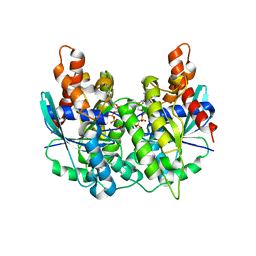 | | Structure of pnob8 ParA | | 分子名称: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ParA | | 著者 | Schumacher, M. | | 登録日 | 2016-05-24 | | 公開日 | 2016-06-22 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (2.369 Å) | | 主引用文献 | Structures of archaeal DNA segregation machinery reveal bacterial and eukaryotic linkages.
Science, 349, 2015
|
|
5K5A
 
 | |
5L1N
 
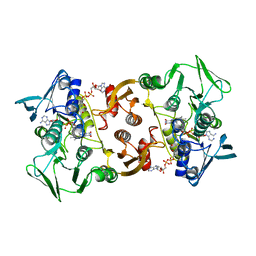 | | Pyrococcus horikoshii CoA Disulfide Reductase Quadruple Mutant | | 分子名称: | COENZYME A, Coenzyme A disulfide reductase, FLAVIN-ADENINE DINUCLEOTIDE | | 著者 | Sea, K, Chen, B, Crane III, E.J, Sazinsky, M.H. | | 登録日 | 2016-07-29 | | 公開日 | 2017-08-09 | | 最終更新日 | 2024-10-09 | | 実験手法 | X-RAY DIFFRACTION (3.6 Å) | | 主引用文献 | A broader active site inPyrococcus horikoshiiCoA disulfide reductase accommodates larger substrates and reveals evidence of subunit asymmetry.
FEBS Open Bio, 8, 2018
|
|
4FJU
 
 | |
4H8A
 
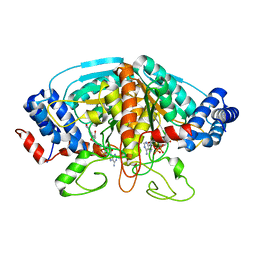 | | Crystal structure of ureidoglycolate dehydrogenase in binary complex with NADH | | 分子名称: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Ureidoglycolate dehydrogenase | | 著者 | Rhee, S, Shin, I, Kim, M. | | 登録日 | 2012-09-22 | | 公開日 | 2013-01-16 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.64 Å) | | 主引用文献 | Structural and functional insights into (s)-ureidoglycolate dehydrogenase, a metabolic branch point enzyme in nitrogen utilization.
Plos One, 7, 2012
|
|
1MRC
 
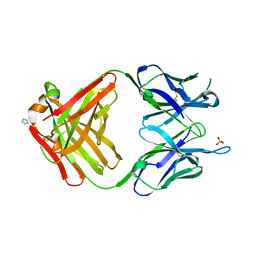 | | PREPARATION, CHARACTERIZATION AND CRYSTALLIZATION OF AN ANTIBODY FAB FRAGMENT THAT RECOGNIZES RNA. CRYSTAL STRUCTURES OF NATIVE FAB AND THREE FAB-MONONUCLEOTIDE COMPLEXES | | 分子名称: | IGG2B-KAPPA JEL103 FAB (HEAVY CHAIN), IGG2B-KAPPA JEL103 FAB (LIGHT CHAIN), IMIDAZOLE, ... | | 著者 | Pokkuluri, P.R, Cygler, M. | | 登録日 | 1994-06-13 | | 公開日 | 1995-02-14 | | 最終更新日 | 2024-06-05 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Preparation, characterization and crystallization of an antibody Fab fragment that recognizes RNA. Crystal structures of native Fab and three Fab-mononucleotide complexes.
J.Mol.Biol., 243, 1994
|
|
1MRD
 
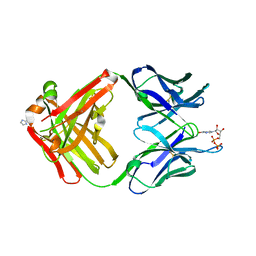 | | PREPARATION, CHARACTERIZATION AND CRYSTALLIZATION OF AN ANTIBODY FAB FRAGMENT THAT RECOGNIZES RNA. CRYSTAL STRUCTURES OF NATIVE FAB AND THREE FAB-MONONUCLEOTIDE COMPLEXES | | 分子名称: | IGG2B-KAPPA JEL103 FAB (HEAVY CHAIN), IGG2B-KAPPA JEL103 FAB (LIGHT CHAIN), IMIDAZOLE, ... | | 著者 | Pokkuluri, P.R, Cygler, M. | | 登録日 | 1994-06-13 | | 公開日 | 1995-02-14 | | 最終更新日 | 2024-06-05 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Preparation, characterization and crystallization of an antibody Fab fragment that recognizes RNA. Crystal structures of native Fab and three Fab-mononucleotide complexes.
J.Mol.Biol., 243, 1994
|
|
1MRE
 
 | | PREPARATION, CHARACTERIZATION AND CRYSTALLIZATION OF AN ANTIBODY FAB FRAGMENT THAT RECOGNIZES RNA. CRYSTAL STRUCTURES OF NATIVE FAB AND THREE FAB-MONONUCLEOTIDE COMPLEXES | | 分子名称: | GUANOSINE-5'-DIPHOSPHATE, IGG2B-KAPPA JEL103 FAB (HEAVY CHAIN), IGG2B-KAPPA JEL103 FAB (LIGHT CHAIN), ... | | 著者 | Pokkuluri, P.R, Cygler, M. | | 登録日 | 1994-06-13 | | 公開日 | 1995-02-14 | | 最終更新日 | 2024-06-05 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Preparation, characterization and crystallization of an antibody Fab fragment that recognizes RNA. Crystal structures of native Fab and three Fab-mononucleotide complexes.
J.Mol.Biol., 243, 1994
|
|
2ESF
 
 | | Identification of a Novel Non-Catalytic Bicarbonate Binding Site in Eubacterial beta-Carbonic Anhydrase | | 分子名称: | BICARBONATE ION, Carbonic anhydrase 2, ZINC ION | | 著者 | Cronk, J.D, Rowlett, R.S, Zhang, K.Y.J, Tu, C, Endrizzi, J.A, Gareiss, P.C. | | 登録日 | 2005-10-26 | | 公開日 | 2006-04-18 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Identification of a Novel Noncatalytic Bicarbonate Binding Site in Eubacterial beta-Carbonic Anhydrase.
Biochemistry, 45, 2006
|
|
5XV9
 
 | |
1OB0
 
 | | Kinetic stabilization of Bacillus licheniformis alpha-amylase through introduction of hydrophobic residues at the surface | | 分子名称: | ALPHA-AMYLASE, CALCIUM ION, SODIUM ION | | 著者 | Machius, M, Declerck, N, Huber, R, Wiegand, G. | | 登録日 | 2003-01-21 | | 公開日 | 2003-01-30 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.83 Å) | | 主引用文献 | Kinetic Stabilization of Bacillus Licheniformis Alpha-Amylase Through Introduction of Hydrophobic Residues at the Surface
J.Biol.Chem., 278, 2003
|
|
