4J19
 
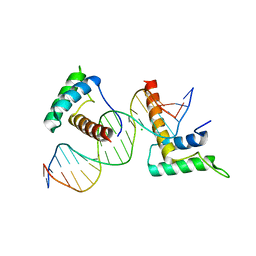 | | Structure of a novel telomere repeat binding protein bound to DNA | | 分子名称: | CHLORIDE ION, DNA (5'-D(*CP*TP*GP*TP*TP*AP*GP*GP*GP*TP*TP*AP*GP*GP*GP*TP*TP*AP*G)-3'), DNA (5'-D(*TP*CP*TP*AP*AP*CP*CP*CP*TP*AP*AP*CP*CP*CP*TP*AP*AP*CP*A)-3'), ... | | 著者 | Kappei, D, Butter, F, Benda, C, Scheibe, M, Draskovic, I, Stevense, M, Lopes Novo, C, Basquin, C, Krastev, D.B, Kittler, R, Jessberger, R, Londono-Vallejo, A.J, Mann, M, Buchholz, F. | | 登録日 | 2013-02-01 | | 公開日 | 2013-05-29 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | HOT1 is a mammalian direct telomere repeat-binding protein contributing to telomerase recruitment.
Embo J., 32, 2013
|
|
7YU1
 
 | |
7YU0
 
 | |
7YU2
 
 | |
1XAB
 
 | |
1XAA
 
 | |
8YXA
 
 | | Crystal structure of the HSA complex with cefazolin and myristate | | 分子名称: | (6~{R},7~{R})-3-[(5-methyl-1,3,4-thiadiazol-2-yl)sulfanylmethyl]-8-oxidanylidene-7-[2-(1,2,3,4-tetrazol-1-yl)ethanoylamino]-5-thia-1-azabicyclo[4.2.0]oct-2-ene-2-carboxylic acid, MYRISTIC ACID, Serum albumin | | 著者 | Kawai, A. | | 登録日 | 2024-04-02 | | 公開日 | 2024-08-07 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Interaction of Cephalosporins with Human Serum Albumin: A Structural Study.
J.Med.Chem., 2024
|
|
8YXB
 
 | |
6IZN
 
 | | Crystal structure of the PPARgamma-LBD complexed with compound 3g | | 分子名称: | 3-[[6-(2,6-dimethylpyridin-3-yl)oxy-7-fluoranyl-1-methyl-benzimidazol-2-yl]methoxy]benzoic acid, Peptide from Peroxisome proliferator-activated receptor gamma coactivator 1-alpha, Peroxisome proliferator-activated receptor gamma | | 著者 | Matsui, Y, Hanzawa, H. | | 登録日 | 2018-12-20 | | 公開日 | 2019-03-27 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Structure-Activity Relationship Studies of 3- or 4-Pyridine Derivatives of DS-6930.
Acs Med.Chem.Lett., 10, 2019
|
|
3FKR
 
 | |
3FKK
 
 | | Structure of L-2-keto-3-deoxyarabonate dehydratase | | 分子名称: | L-2-keto-3-deoxyarabonate dehydratase, PHOSPHATE ION | | 著者 | Shimada, N, Mikami, B. | | 登録日 | 2008-12-17 | | 公開日 | 2010-01-12 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structural analysis of L -2-keto-3-deoxyarabonate dehydratase an enzyme involved in an alternative bacterial pathway of L-arabinose metabolism in complex with pyruvate
To be Published
|
|
3ABQ
 
 | |
3ABR
 
 | |
3ABO
 
 | |
3ABS
 
 | |
6IZM
 
 | | Crystal structure of the PPARgamma-LBD complexed with compound 1l | | 分子名称: | 3-[[6-(2,6-dimethylpyridin-3-yl)oxy-1-methyl-benzimidazol-2-yl]methoxy]benzoic acid, Peptide from Peroxisome proliferator-activated receptor gamma coactivator 1-alpha, Peroxisome proliferator-activated receptor gamma | | 著者 | Matsui, Y, Hanzawa, H. | | 登録日 | 2018-12-20 | | 公開日 | 2019-03-27 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structure-Activity Relationship Studies of 3- or 4-Pyridine Derivatives of DS-6930.
Acs Med.Chem.Lett., 10, 2019
|
|
4NXS
 
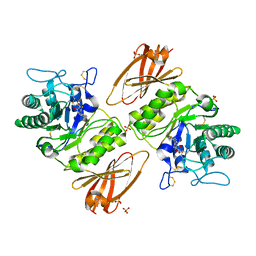 | | Crystal structure of human alpha-galactosidase A in complex with 1-deoxygalactonojirimycin-pFPhT | | 分子名称: | (2R,3S,4R,5S)-N-(4-fluorophenyl)-3,4,5-trihydroxy-2-(hydroxymethyl)piperidine-1-carbothioamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Johnson, J.L, Drury, J.E, Lieberman, R.L. | | 登録日 | 2013-12-09 | | 公開日 | 2014-06-11 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.5493 Å) | | 主引用文献 | Molecular Basis of 1-Deoxygalactonojirimycin Arylthiourea Binding to Human alpha-Galactosidase A: Pharmacological Chaperoning Efficacy on Fabry Disease Mutants.
Acs Chem.Biol., 9, 2014
|
|
1GD6
 
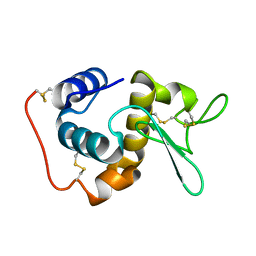 | | STRUCTURE OF THE BOMBYX MORI LYSOZYME | | 分子名称: | LYSOZYME | | 著者 | Matsuura, A, Aizawa, T, Yao, M, Kawano, K, Tanaka, I, Nitta, K. | | 登録日 | 2000-09-19 | | 公開日 | 2001-03-21 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structural analysis of an insect lysozyme exhibiting catalytic efficiency at low temperatures.
Biochemistry, 41, 2002
|
|
5B5F
 
 | | Crystal structure of ALiS3-Streptavidin complex | | 分子名称: | N-methyl-3-(4-oxo-4,5-dihydrofuro[3,2-c]pyridin-2-yl)benzenesulfonamide, Streptavidin | | 著者 | Sugiyama, S, Terai, T, Kakinouchi, K, Fujikake, R, Nagano, T, Urano, Y. | | 登録日 | 2016-05-04 | | 公開日 | 2017-03-01 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.2 Å) | | 主引用文献 | Improving the Solubility of Artificial Ligands of Streptavidin to Enable More Practical Reversible Switching of Protein Localization in Cells
Chembiochem, 18, 2017
|
|
5B6F
 
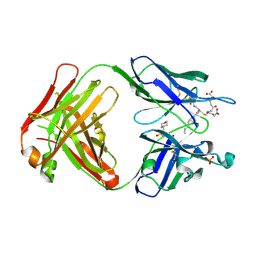 | | Crystal structure of the Fab fragment of an anti-Leukotriene C4 monoclonal antibody complexed with LTC4 | | 分子名称: | (5~{S},6~{R},7~{E},9~{E},11~{Z},14~{Z})-6-[(2~{R})-2-[[(4~{S})-4-azanyl-5-oxidanyl-5-oxidanylidene-pentanoyl]amino]-3-(2-hydroxy-2-oxoethylamino)-3-oxidanylidene-propyl]sulfanyl-5-oxidanyl-icosa-7,9,11,14-tetraenoic acid, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, SULFATE ION, ... | | 著者 | Sugahara, M, Ago, H, Saino, H, Miyano, M. | | 登録日 | 2016-05-27 | | 公開日 | 2017-05-31 | | 最終更新日 | 2019-07-24 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Crystal structure of the Fab fragment of an anti-Leukotriene C4 monoclonal antibody complexed with LTC4
To Be Published
|
|
6VXQ
 
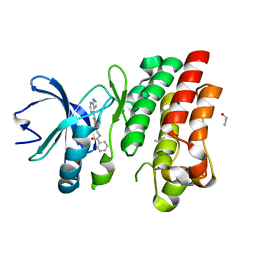 | | Bruton's tyrosine kinase in complex with compound 5 | | 分子名称: | DIMETHYL SULFOXIDE, N-{[4-(7H-pyrrolo[2,3-d]pyrimidin-4-yl)phenyl]methyl}benzamide, Tyrosine-protein kinase BTK | | 著者 | Metrick, C.M, Marcotte, D.J. | | 登録日 | 2020-02-23 | | 公開日 | 2020-12-16 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Discovery of BIIB068: A Selective, Potent, Reversible Bruton's Tyrosine Kinase Inhibitor as an Orally Efficacious Agent for Autoimmune Diseases.
J.Med.Chem., 63, 2020
|
|
6W06
 
 | | Bruton's tyrosine kinase in complex with compound 6 | | 分子名称: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 4-~{tert}-butyl-~{N}-[[4-(7~{H}-pyrrolo[2,3-d]pyrimidin-4-yl)phenyl]methyl]benzamide, DIMETHYL SULFOXIDE, ... | | 著者 | Metrick, C.M, Marcotte, D.J. | | 登録日 | 2020-02-29 | | 公開日 | 2020-12-16 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Discovery of BIIB068: A Selective, Potent, Reversible Bruton's Tyrosine Kinase Inhibitor as an Orally Efficacious Agent for Autoimmune Diseases.
J.Med.Chem., 63, 2020
|
|
1IYK
 
 | | Crystal structure of candida albicans N-myristoyltransferase with myristoyl-COA and peptidic inhibitor | | 分子名称: | MYRISTOYL-COA:PROTEIN N-MYRISTOYLTRANSFERASE, TETRADECANOYL-COA, [CYCLOHEXYLETHYL]-[[[[4-[2-METHYL-1-IMIDAZOLYL-BUTYL]PHENYL]ACETYL]-SERYL]-LYSINYL]-AMINE | | 著者 | Sogabe, S, Fukami, T.A, Morikami, K, Shiratori, Y, Aoki, Y, D'Arcy, A, Winkler, F.K, Banner, D.W, Ohtsuka, T. | | 登録日 | 2002-08-29 | | 公開日 | 2002-12-30 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Crystal Structures of Candida albicans N-Myristoyltransferase with Two Distinct Inhibitors
CHEM.BIOL., 9, 2002
|
|
5GOW
 
 | |
7X7X
 
 | | Human serum albumin complex with deschloro-aripiprazole | | 分子名称: | 7-[4-(4-phenylpiperazin-1-yl)butoxy]-3,4-dihydro-1H-quinolin-2-one, PHOSPHATE ION, Serum albumin | | 著者 | Kawai, A, Otagiri, M. | | 登録日 | 2022-03-10 | | 公開日 | 2022-09-07 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Chlorine Atoms of an Aripiprazole Molecule Control the Geometry and Motion of Aripiprazole and Deschloro-aripiprazole in Subdomain IIIA of Human Serum Albumin.
Acs Omega, 7, 2022
|
|
