5DN1
 
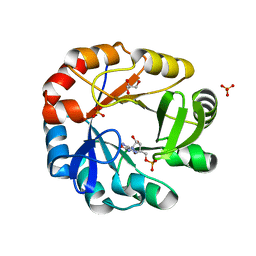 | | Crystal structure of Phosphoribosyl isomerase A from Streptomyces coelicolor | | 分子名称: | AMINOIMIDAZOLE 4-CARBOXAMIDE RIBONUCLEOTIDE, GLYCEROL, Phosphoribosyl isomerase A, ... | | 著者 | Chang, C, Verduzco-Castro, E.A, Endres, M, Barona-Gomez, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2015-09-09 | | 公開日 | 2015-09-30 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.953 Å) | | 主引用文献 | Co-occurrence of analogous enzymes determines evolution of a novel ( beta alpha )8-isomerase sub-family after non-conserved mutations in flexible loop.
Biochem. J., 473, 2016
|
|
5E2H
 
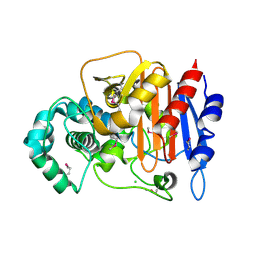 | | Crystal Structure of D-alanine Carboxypeptidase AmpC from Mycobacterium smegmatis | | 分子名称: | Beta-lactamase, CHLORIDE ION, GLYCEROL | | 著者 | Kim, Y, Hatzos-Skintges, C, Endres, M, Babnigg, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2015-10-01 | | 公開日 | 2015-10-14 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal Structure of D-alanine Carboxypeptidase AmpC from Mycobacterium smegmatis
To Be Published
|
|
5E43
 
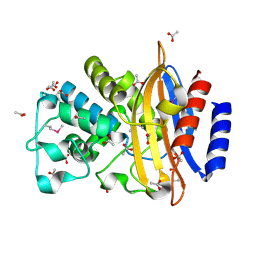 | | Crystal Structure of Beta-lactamase Sros_5706 from Streptosporangium roseum | | 分子名称: | ACETATE ION, Beta-lactamase, NITRATE ION | | 著者 | Kim, Y, Hatzos-Skintges, C, Endres, M, Babnigg, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2015-10-05 | | 公開日 | 2015-10-14 | | 最終更新日 | 2019-12-25 | | 実験手法 | X-RAY DIFFRACTION (1.7095 Å) | | 主引用文献 | Crystal Structure of Beta-lactamase Sros_5706 from Streptosporangium roseum
To Be Published
|
|
5CR9
 
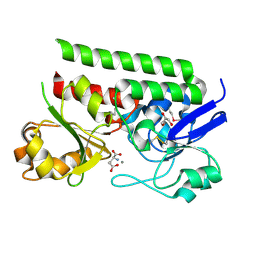 | | Crystal structure of ABC-type Fe3+-hydroxamate transport system from Saccharomonospora viridis DSM 43017 | | 分子名称: | ABC-type Fe3+-hydroxamate transport system, periplasmic component, GLUTAMIC ACID, ... | | 著者 | Nocek, B, Cuff, M, Mack, J, Endres, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2015-07-22 | | 公開日 | 2015-08-12 | | 最終更新日 | 2019-12-25 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Crystal structure of ABC-type Fe3+-hydroxamate transport system from Saccharomonospora viridis DSM 43017
To Be Published
|
|
5DD4
 
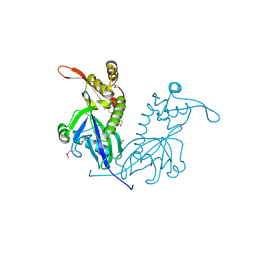 | | Apo structure of transcriptional factor AraR from Bacteroides thetaiotaomicron VPI | | 分子名称: | 1,2-ETHANEDIOL, ACETATE ION, TRANSCRIPTIONAL REGULATOR AraR | | 著者 | Chang, C, Tesar, C, Rodionov, D, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2015-08-24 | | 公開日 | 2015-09-09 | | 最終更新日 | 2015-12-16 | | 実験手法 | X-RAY DIFFRACTION (2.56 Å) | | 主引用文献 | A novel transcriptional regulator of L-arabinose utilization in human gut bacteria.
Nucleic Acids Res., 43, 2015
|
|
5CXW
 
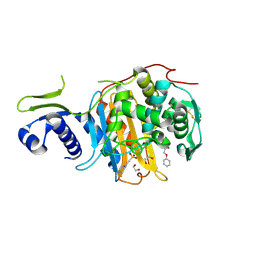 | | Structure of the PonA1 protein from Mycobacterium Tuberculosis in complex with penicillin V | | 分子名称: | (2R,4S)-5,5-dimethyl-2-{(1R)-2-oxo-1-[(phenoxyacetyl)amino]ethyl}-1,3-thiazolidine-4-carboxylic acid, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Filippova, E.V, Kiryukhina, O, Kieser, K, Endres, M, Rubin, E, Sacchettini, J, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG), Structures of Mtb Proteins Conferring Susceptibility to Known Mtb Inhibitors (MTBI) | | 登録日 | 2015-07-29 | | 公開日 | 2016-05-04 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Crystal structures of the transpeptidase domain of the Mycobacterium tuberculosis penicillin-binding protein PonA1 reveal potential mechanisms of antibiotic resistance.
Febs J., 283, 2016
|
|
5EVL
 
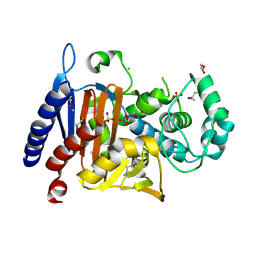 | | Crystal Structure of Beta-Lactamase/D-Alanine Carboxypeptidase from Chromobacterium violaceum | | 分子名称: | Beta-lactamase, CHLORIDE ION, GLYCEROL, ... | | 著者 | Kim, Y, Hatzos-Skintges, C, Endres, M, Babnigg, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2015-11-19 | | 公開日 | 2015-12-02 | | 最終更新日 | 2019-11-27 | | 実験手法 | X-RAY DIFFRACTION (1.52 Å) | | 主引用文献 | Crystal Structure of Beta-Lactamase/D-Alanine Carboxypeptidase from Chromobacterium violaceum
To Be Published
|
|
5F1P
 
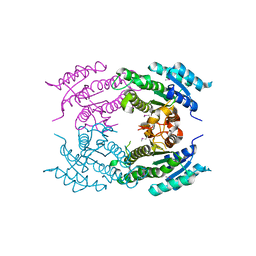 | | Crystal Structure of Dehydrogenase from Streptomyces platensis | | 分子名称: | PtmO8 | | 著者 | Kim, Y, Li, H, Endres, M, Babnigg, G, Rudolf, J, Ma, M, Chang, C.-Y, Shen, B, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | 登録日 | 2015-11-30 | | 公開日 | 2015-12-30 | | 最終更新日 | 2019-12-04 | | 実験手法 | X-RAY DIFFRACTION (2.099 Å) | | 主引用文献 | Crystal Structure of a Dehydrogenase, PtmO8, from Streptomyces platensis
To Be Published
|
|
5F4B
 
 | | Structure of B. abortus WrbA-related protein A (WrpA) | | 分子名称: | CHLORIDE ION, FLAVIN MONONUCLEOTIDE, NAD(P)H dehydrogenase (quinone) | | 著者 | Herrou, J, Czyz, D, Willett, J.W, Kim, H.S, Chhor, G, Endres, M, Babnigg, G, Kim, Y, Joachimiak, A, Crosson, S, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2015-12-03 | | 公開日 | 2016-03-09 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (2.498 Å) | | 主引用文献 | WrpA Is an Atypical Flavodoxin Family Protein under Regulatory Control of the Brucella abortus General Stress Response System.
J.Bacteriol., 198, 2016
|
|
1M6Y
 
 | | Crystal Structure Analysis of TM0872, a Putative SAM-dependent Methyltransferase, Complexed with SAH | | 分子名称: | S-ADENOSYL-L-HOMOCYSTEINE, S-adenosyl-methyltransferase mraW, SULFATE ION | | 著者 | Miller, D.J, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2002-07-17 | | 公開日 | 2003-01-28 | | 最終更新日 | 2016-03-30 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Crystal complexes of a predicted S-adenosylmethionine-dependent methyltransferase reveal a typical AdoMet binding domain and a substrate recognition domain
Protein Sci., 12, 2003
|
|
1LJ9
 
 | | The crystal structure of the transcriptional regulator SlyA | | 分子名称: | transcriptional regulator SlyA | | 著者 | Wu, R.Y, Zhang, R.G, Gornicki, P, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2002-04-19 | | 公開日 | 2003-01-21 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Crystal structure of Enterococcus faecalis SlyA-like transcriptional factor
J.Biol.Chem., 278, 2003
|
|
1N2X
 
 | | Crystal Structure Analysis of TM0872, a Putative SAM-dependent Methyltransferase, Complexed with SAM | | 分子名称: | S-ADENOSYLMETHIONINE, S-adenosyl-methyltransferase mraW, SULFATE ION | | 著者 | Miller, D.J, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2002-10-24 | | 公開日 | 2003-01-28 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Crystal complexes of a predicted S-adenosylmethionine-dependent methyltransferase reveal a typical AdoMet binding domain and a substrate recognition domain
Protein Sci., 12, 2003
|
|
3VDH
 
 | | Crystal structure of PbGH5A, a glycoside hydrolase family 5 enzyme from Prevotella bryantii B14 | | 分子名称: | B-1,4-endoglucanase, CHLORIDE ION | | 著者 | Stogios, P.J, Evdokimova, E, Egorova, O, Yim, V, Joachimiak, A, Edwards, A.M, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2012-01-05 | | 公開日 | 2012-01-18 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (1.62 Å) | | 主引用文献 | Structure-Function Analysis of a Mixed-linkage beta-Glucanase/Xyloglucanase from the Key Ruminal Bacteroidetes Prevotella bryantii B14.
J.Biol.Chem., 291, 2016
|
|
3VCX
 
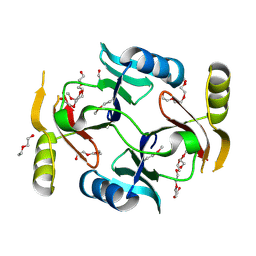 | | Crystal structure of a putative glyoxalase/bleomycin resistance protein from Rhodopseudomonas palustris CGA009 | | 分子名称: | Glyoxalase/Bleomycin resistance protein/dioxygenase domain, TETRAETHYLENE GLYCOL | | 著者 | Stogios, P.J, Chang, C, Evdokimova, E, Egorova, O, Joachimiak, A, Edwards, A.M, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2012-01-04 | | 公開日 | 2012-01-18 | | 最終更新日 | 2012-01-25 | | 実験手法 | X-RAY DIFFRACTION (1.39 Å) | | 主引用文献 | Crystal structure of a putative glyoxalase/bleomycin resistance protein from Rhodopseudomonas palustris CGA009
To be Published
|
|
3B7H
 
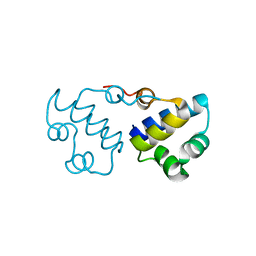 | |
3B4S
 
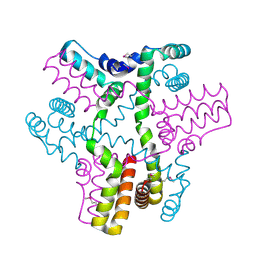 | |
3B49
 
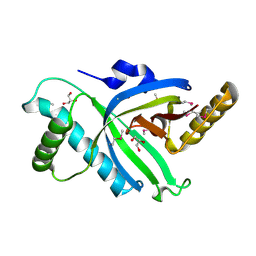 | |
3B4Q
 
 | |
3B85
 
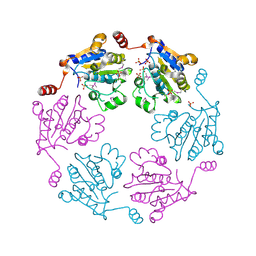 | |
3B79
 
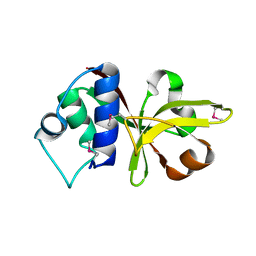 | |
3BJD
 
 | | Crystal structure of putative 3-oxoacyl-(acyl-carrier-protein) synthase from Pseudomonas aeruginosa | | 分子名称: | 1,2-ETHANEDIOL, NICKEL (II) ION, Putative 3-oxoacyl-(acyl-carrier-protein) synthase | | 著者 | Osipiuk, J, Evdokimova, E, Kudritska, M, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2007-12-03 | | 公開日 | 2007-12-11 | | 最終更新日 | 2017-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Crystal structure of putative 3-oxoacyl-(acyl-carrier-protein) synthase from Pseudomonas aeruginosa.
To be Published
|
|
3B33
 
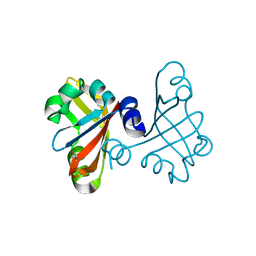 | |
3BJN
 
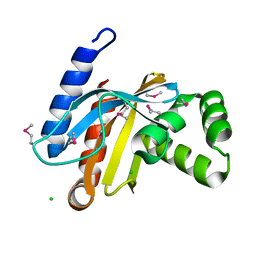 | | Crystal structure of C-terminal domain of putative transcriptional regulator from Vibrio cholerae, targeted domain 79-240 | | 分子名称: | CHLORIDE ION, Transcriptional regulator, putative | | 著者 | Chang, C, Volkart, L, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2007-12-04 | | 公開日 | 2007-12-11 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Crystal structure of C-terminal domain of putative transcriptional regulator from Vibrio cholerae.
To be Published
|
|
3B8F
 
 | | Crystal structure of the cytidine deaminase from Bacillus anthracis | | 分子名称: | Putative Blasticidin S deaminase | | 著者 | Zhang, R, Joachimiak, G, Wu, R, Patterson, S, Gornicki, P, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2007-11-01 | | 公開日 | 2007-12-04 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | The crystal structure of the cytidine deaminase from Bacillus anthracis.
To be Published
|
|
3BEE
 
 | | Crystal structure of putative YfrE protein from Vibrio parahaemolyticus | | 分子名称: | 1,2-ETHANEDIOL, Putative YfrE protein | | 著者 | Chang, C, Hatzos, C, Abdullah, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2007-11-16 | | 公開日 | 2007-12-04 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | Crystal structure of putative YfrE protein from Vibrio parahaemolyticus.
To be Published
|
|
