1YEZ
 
 | | Solution structure of the conserved protein from the gene locus MM1357 of Methanosarcina mazei. Northeast Structural Genomics target MaR30. | | 分子名称: | MM1357 | | 著者 | Rossi, P, Aramini, J.M, Swapna, G.V.T, Huang, Y.P, Xiao, R, Ho, C.K, Ma, L.C, Acton, T.B, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2004-12-29 | | 公開日 | 2005-02-22 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of the conserved protein from the gene locus MM1357 of Methanosarcina mazei. Northeast Structural Genomics target MaR30.
To be Published
|
|
1YRS
 
 | | Crystal structure of KSP in complex with inhibitor 1 | | 分子名称: | 3-[(5S)-1-ACETYL-3-(2-CHLOROPHENYL)-4,5-DIHYDRO-1H-PYRAZOL-5-YL]PHENOL, ADENOSINE-5'-DIPHOSPHATE, Kinesin-like protein KIF11, ... | | 著者 | Cox, C.D, Breslin, M.J, Mariano, B.J, Coleman, P.J, Buser, C.A, Walsh, E.S, Hamilton, K, Kohl, N.E, Torrent, M, Yan, Y, Kuo, L.C, Hartman, G.D. | | 登録日 | 2005-02-04 | | 公開日 | 2005-04-12 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Kinesin spindle protein (KSP) inhibitors. Part 1: The discovery of 3,5-diaryl-4,5-dihydropyrazoles as potent and selective inhibitors of the mitotic kinesin KSP
BIOORG.MED.CHEM.LETT., 15, 2005
|
|
1LL9
 
 | | Crystal Structure Of AmpC beta-Lactamase From E. Coli In Complex With Amoxicillin | | 分子名称: | 2-{1-[2-AMINO-2-(4-HYDROXY-PHENYL)-ACETYLAMINO]-2-OXO-ETHYL}-5,5-DIMETHYL-THIAZOLIDINE-4-CARBOXYLIC ACID, beta-lactamase | | 著者 | Trehan, I, Morandi, F, Blaszczak, L.C, Shoichet, B.K. | | 登録日 | 2002-04-26 | | 公開日 | 2002-10-02 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (1.87 Å) | | 主引用文献 | Using steric hindrance to design new inhibitors of class C beta-lactamases.
Chem.Biol., 9, 2002
|
|
1XW3
 
 | | Crystal Structure of Human Sulfiredoxin (Srx) | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, PHOSPHATE ION, SULFIREDOXIN | | 著者 | Murray, M.S, Jonsson, T.J, Johnson, L.C, Poole, L.B, Lowther, W.T. | | 登録日 | 2004-10-29 | | 公開日 | 2005-05-24 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Structural basis for the retroreduction of inactivated peroxiredoxins by human sulfiredoxin.
Biochemistry, 44, 2005
|
|
1KXS
 
 | |
1KWS
 
 | | CRYSTAL STRUCTURE OF BETA1,3-GLUCURONYLTRANSFERASE I IN COMPLEX WITH THE ACTIVE UDP-GLCUA DONOR | | 分子名称: | BETA-1,3-GLUCURONYLTRANSFERASE 3, MANGANESE (II) ION, URIDINE-5'-DIPHOSPHATE-GLUCURONIC ACID | | 著者 | Pedersen, L.C, Darden, T.A, Negishi, M. | | 登録日 | 2002-01-30 | | 公開日 | 2002-06-19 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Crystal structure of beta 1,3-glucuronyltransferase I in complex with active donor substrate UDP-GlcUA.
J.Biol.Chem., 277, 2002
|
|
1LLB
 
 | | Crystal Structure Of AmpC beta-Lactamase From E. Coli In Complex With ATMO-penicillin | | 分子名称: | 2-{1-[2-(2-AMINO-THIAZOL-4-YL)-2-METHOXYIMINO-ACETYLAMINO]-2-OXO-ETHYL}-5,5-DIMETHYL-THIAZOLIDINE-4-CARBOXYLIC ACID, beta-lactamase | | 著者 | Trehan, I, Morandi, F, Blaszczak, L.C, Shoichet, B.K. | | 登録日 | 2002-04-26 | | 公開日 | 2002-10-02 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (1.72 Å) | | 主引用文献 | Using steric hindrance to design new inhibitors of class C beta-lactamases.
Chem.Biol., 9, 2002
|
|
1XDB
 
 | | Crystal Structure of the Nitrogenase Fe protein Asp129Glu | | 分子名称: | IRON/SULFUR CLUSTER, Nitrogenase iron protein 1 | | 著者 | Jang, S.B, Jeong, M.S, Seefeldt, L.C, Peters, J.W. | | 登録日 | 2004-09-05 | | 公開日 | 2005-03-01 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Structural and biochemical implications of single amino acid substitutions in the nucleotide-dependent switch regions of the nitrogenase Fe protein from Azotobacter vinelandii
J.Biol.Inorg.Chem., 9, 2004
|
|
1L6O
 
 | | XENOPUS DISHEVELLED PDZ DOMAIN | | 分子名称: | Dapper 1, Segment polarity protein dishevelled homolog DVL-2 | | 著者 | Cheyette, B.N.R, Waxman, J.S, Miller, J.R, Takemaru, K.-I, Sheldahl, L.C, Khlebtsova, N, Fox, E.P, Earnest, T, Moon, R.T. | | 登録日 | 2002-03-11 | | 公開日 | 2003-06-03 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Dapper, a Dishevelled-associated antagonist of beta-catenin and JNK signaling, is required for notochord formation
Dev.Cell, 2, 2002
|
|
1XPW
 
 | | Solution NMR Structure of human protein HSPCO34. Northeast Structural Genomics Target HR1958 | | 分子名称: | LOC51668 protein | | 著者 | Ramelot, T.A, Xiao, R, Ma, L.C, Acton, T.B, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2004-10-09 | | 公開日 | 2004-11-09 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Improving NMR protein structure quality by Rosetta refinement: a molecular replacement study.
Proteins, 75, 2009
|
|
1DUR
 
 | |
1ZGV
 
 | | Thrombin in complex with an oxazolopyridine inhibitor 2 | | 分子名称: | Hirudin, N7-BUTYL-N2-(5-CHLORO-2-METHYLPHENYL)-5-METHYL[1,2,4]TRIAZOLO[1,5-A]PYRIMIDINE-2,7-DIAMINE, Thrombin | | 著者 | Deng, J.Z, McMasters, D.R, Rabbat, P.M, Williams, P.D, Coburn, C.A, Yan, Y, Kuo, L.C, Lewis, S.D, Lucas, B.J, Krueger, J.A, Strulovici, B, Vacca, J.P, Lyle, T.A, Burgey, C.S. | | 登録日 | 2005-04-22 | | 公開日 | 2005-09-27 | | 最終更新日 | 2013-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Development of an oxazolopyridine series of dual thrombin/factor Xa inhibitors via structure-guided lead optimization.
Bioorg.Med.Chem.Lett., 15, 2005
|
|
1ZM1
 
 | | Crystal structures of complex F. succinogenes 1,3-1,4-beta-D-glucanase and beta-1,3-1,4-cellotriose | | 分子名称: | Beta-glucanase, CALCIUM ION, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose | | 著者 | Tsai, L.C, Shyur, L.F, Cheng, Y.S, Lee, S.H. | | 登録日 | 2005-05-10 | | 公開日 | 2006-05-10 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Crystal structure of truncated Fibrobacter succinogenes 1,3-1,4-beta-D-glucanase in complex with beta-1,3-1,4-cellotriose
J.Mol.Biol., 354, 2005
|
|
2AAK
 
 | |
1M5A
 
 | |
5GNY
 
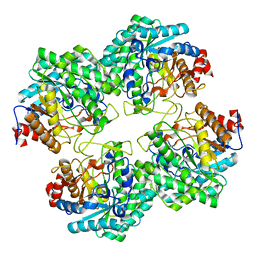 | | The structure of WT Bgl6 | | 分子名称: | Beta-glucosidase, beta-D-glucopyranose | | 著者 | Xie, W, Pang, P, Cao, L.C, Liu, Y.H, Wang, Z. | | 登録日 | 2016-07-25 | | 公開日 | 2017-04-12 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.105 Å) | | 主引用文献 | Structures of a glucose-tolerant beta-glucosidase provide insights into its mechanism.
J. Struct. Biol., 198, 2017
|
|
5GNX
 
 | | The E171Q mutant structure of Bgl6 | | 分子名称: | Beta-glucosidase, GLYCEROL, PROPANOIC ACID, ... | | 著者 | Xie, W, Pang, P, Cao, L.C, Liu, Y.H, Wang, Z. | | 登録日 | 2016-07-25 | | 公開日 | 2017-04-12 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structures of a glucose-tolerant beta-glucosidase provide insights into its mechanism.
J. Struct. Biol., 198, 2017
|
|
5GNZ
 
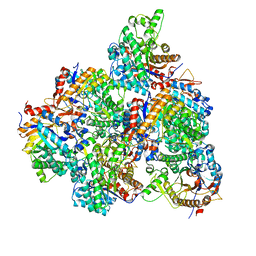 | | The M3 mutant structure of Bgl6 | | 分子名称: | Beta-glucosidase, GLYCEROL, beta-D-glucopyranose | | 著者 | Xie, W, Pang, P, Cao, L.C, Liu, Y.H, Wang, Z. | | 登録日 | 2016-07-25 | | 公開日 | 2017-04-12 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Structures of a glucose-tolerant beta-glucosidase provide insights into its mechanism.
J. Struct. Biol., 198, 2017
|
|
5HGP
 
 | |
5HGL
 
 | | Hexameric HIV-1 CA, open conformation | | 分子名称: | CHLORIDE ION, Capsid protein P24, N-METHYL-NALPHA-[(2-METHYL-1H-INDOL-3-YL)ACETYL]-N-PHENYL-L-PHENYLALANINAMIDE | | 著者 | Price, A.J, Jacques, D.A, James, L.C. | | 登録日 | 2016-01-08 | | 公開日 | 2016-08-10 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | HIV-1 uses dynamic capsid pores to import nucleotides and fuel encapsidated DNA synthesis.
Nature, 536, 2016
|
|
5HGK
 
 | |
5HGO
 
 | |
5HGM
 
 | |
5CO6
 
 | |
3H4L
 
 | | Crystal Structure of N terminal domain of a DNA repair protein | | 分子名称: | DNA mismatch repair protein PMS1, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | 著者 | Arana, M.E, Holmes, S.F, Fortune, J.M, Moon, A.F, Pedersen, L.C, Kunkel, T.A. | | 登録日 | 2009-04-20 | | 公開日 | 2010-03-02 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Functional residues on the surface of the N-terminal domain of yeast Pms1.
Dna Repair, 9, 2010
|
|
