7GCC
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with ALP-POS-95b75b4d-4 (Mpro-x10566) | | 分子名称: | 2-(3-cyclopropylphenyl)-N-(4-methylpyridin-3-yl)acetamide, 3C-like proteinase, DIMETHYL SULFOXIDE | | 著者 | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | 登録日 | 2023-08-11 | | 公開日 | 2023-11-08 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (1.61 Å) | | 主引用文献 | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
3JU1
 
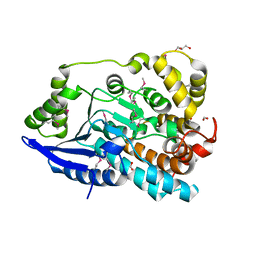 | | Crystal Structure of Enoyl-CoA Hydratase/Isomerase Family Protein | | 分子名称: | ACETIC ACID, Enoyl-CoA hydratase/isomerase family protein, FORMIC ACID, ... | | 著者 | Kim, Y, Xu, X, Cui, H, Ng, J, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2009-09-14 | | 公開日 | 2009-09-22 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2.301 Å) | | 主引用文献 | Crystal Structure of Enoyl-CoA Hydratase/Isomerase Family Protein
To be Published
|
|
7GH1
 
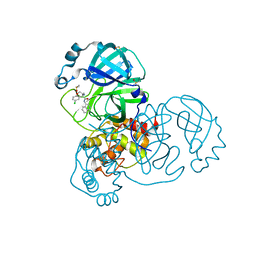 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with MAT-POS-2bb0cf2b-1 (Mpro-x12731) | | 分子名称: | (4S)-4-(aminomethyl)-6-chloro-N-(isoquinolin-4-yl)-3,4-dihydro-2H-1-benzopyran-4-carboxamide, 3C-like proteinase, DIMETHYL SULFOXIDE | | 著者 | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | 登録日 | 2023-08-11 | | 公開日 | 2023-11-08 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (1.52 Å) | | 主引用文献 | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
7GCT
 
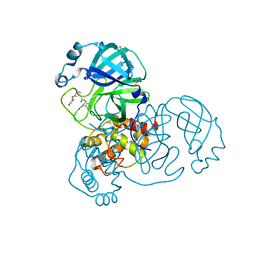 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with BRU-LEF-c49414a7-1 (Mpro-x10756) | | 分子名称: | 3C-like proteinase, DIMETHYL SULFOXIDE, N-(4-methylpyridin-3-yl)-2-(3-{[(2R)-4-oxoazetidin-2-yl]oxy}phenyl)acetamide | | 著者 | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | 登録日 | 2023-08-11 | | 公開日 | 2023-11-08 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (1.857 Å) | | 主引用文献 | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
7GH7
 
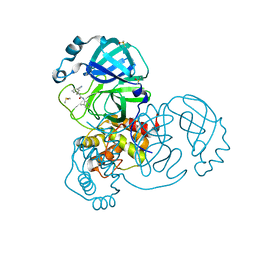 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with TRY-UNI-714-22 (Mpro-x2912) | | 分子名称: | (2R)-2-(3-cyanophenyl)-N-(4-methylpyridin-3-yl)propanamide, 3C-like proteinase, DIMETHYL SULFOXIDE | | 著者 | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | 登録日 | 2023-08-11 | | 公開日 | 2023-11-08 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (1.67 Å) | | 主引用文献 | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
7GD1
 
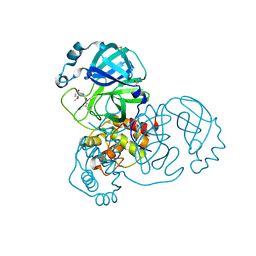 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with BAR-COM-0f94fc3d-58 (Mpro-x10856) | | 分子名称: | (2R)-2-amino-2-(5-bromo-2-methoxyphenyl)-N-(4-methylpyridin-3-yl)acetamide, 3C-like proteinase, DIMETHYL SULFOXIDE | | 著者 | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | 登録日 | 2023-08-11 | | 公開日 | 2023-11-08 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (1.52 Å) | | 主引用文献 | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
7EVV
 
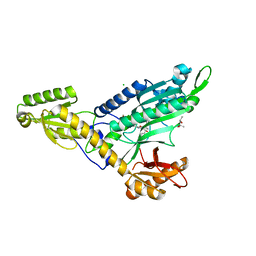 | | Co-crystal Structure of Toxoplasma gondii Prolyl tRNA Synthetase (TgPRS) in complex with JE6 and L-pro | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, PROLINE, ... | | 著者 | Malhotra, N, Mishra, S, Manickam, Y, Sharma, A. | | 登録日 | 2021-05-22 | | 公開日 | 2022-03-09 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.704 Å) | | 主引用文献 | Double drugging of prolyl-tRNA synthetase provides a new paradigm for anti-infective drug development.
Plos Pathog., 18, 2022
|
|
3KYE
 
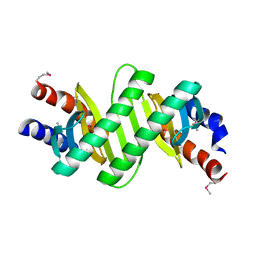 | | Crystal Structure of Roadblock/LC7 Domain from Streptomyces avermitilis | | 分子名称: | Roadblock/LC7 domain, Robl_LC7 | | 著者 | Kim, Y, Xu, X, Cui, H, Ng, J, Edwards, A, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2009-12-05 | | 公開日 | 2009-12-22 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | Crystal Structure of Roadblock/LC7 Domain from Streptomyces avermitilis
To be Published
|
|
3KZV
 
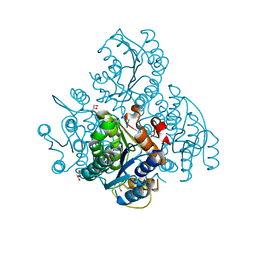 | | The crystal structure of a cytoplasmic protein with unknown function from Saccharomyces cerevisiae | | 分子名称: | GLYCEROL, Uncharacterized oxidoreductase YIR035C | | 著者 | Zhang, R, Xu, X, Cui, H, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2009-12-08 | | 公開日 | 2010-03-02 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | The crystal structure of a cytoplasmic protein with unknown function from Saccharomyces cerevisiae
To be Published
|
|
1ASQ
 
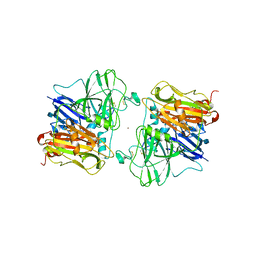 | | X-RAY STRUCTURES AND MECHANISTIC IMPLICATIONS OF THREE FUNCTIONAL DERIVATIVES OF ASCORBATE OXIDASE FROM ZUCCHINI: REDUCED-, PEROXIDE-, AND AZIDE-FORMS | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ASCORBATE OXIDASE, AZIDE ION, ... | | 著者 | Messerschmidt, A, Luecke, H, Huber, R. | | 登録日 | 1992-11-25 | | 公開日 | 1994-01-31 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (2.32 Å) | | 主引用文献 | X-ray structures and mechanistic implications of three functional derivatives of ascorbate oxidase from zucchini. Reduced, peroxide and azide forms.
J.Mol.Biol., 230, 1993
|
|
6C1H
 
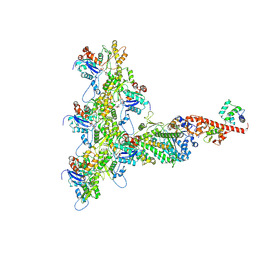 | | High-Resolution Cryo-EM Structures of Actin-bound Myosin States Reveal the Mechanism of Myosin Force Sensing | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | 著者 | Mentes, A, Huehn, A, Liu, X, Zwolak, A, Dominguez, R, Shuman, H, Ostap, E.M, Sindelar, C.V. | | 登録日 | 2018-01-04 | | 公開日 | 2018-01-31 | | 最終更新日 | 2024-03-13 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | High-resolution cryo-EM structures of actin-bound myosin states reveal the mechanism of myosin force sensing.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
3L19
 
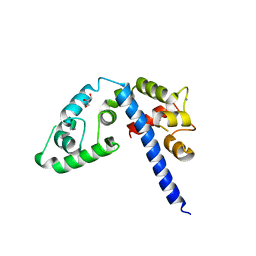 | | Crystal structure of calcium binding domain of CpCDPK3, cgd5_820 | | 分子名称: | CALCIUM ION, Calcium/calmodulin dependent protein kinase with a kinase domain and 4 calmodulin like EF hands, GLYCEROL, ... | | 著者 | Qiu, W, Hutchinson, A, Wernimont, A, Walker, J.R, Sullivan, H, Lin, Y.-H, Mackenzie, F, Kozieradzki, I, Cossar, D, Schapira, M, Senisterra, G, Vedadi, M, Arrowsmith, C.H, Bountra, C, Weigelt, J, Edwards, A.M, Bochkarev, A, Hui, R, Amani, M, Structural Genomics Consortium (SGC) | | 登録日 | 2009-12-11 | | 公開日 | 2009-12-22 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.14 Å) | | 主引用文献 | Crystal structure of calcium binding domain of CpCDPK3, cgd5_820
To be Published
|
|
3KXR
 
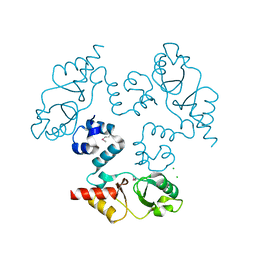 | | Structure of the cystathionine beta-synthase pair domain of the putative Mg2+ transporter SO5017 from Shewanella oneidensis MR-1. | | 分子名称: | CHLORIDE ION, Magnesium transporter, putative | | 著者 | Fratczak, Z, Zimmerman, M.D, Chruszcz, M, Cymborowski, M, Kagan, O, Savchenko, A, Edwards, A, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2009-12-03 | | 公開日 | 2009-12-15 | | 最終更新日 | 2022-04-13 | | 実験手法 | X-RAY DIFFRACTION (2.41 Å) | | 主引用文献 | Structure of the cystathionine beta-synthase pair domain of the putative Mg2+ transporter SO5017 from Shewanella oneidensis MR-1.
To be Published
|
|
5WAH
 
 | | SOLUTION NMR STRUCTURE OF SIGLEC-5 BINDING DOMAIN FROM STREPTOCOCCAL BETA PROTEIN | | 分子名称: | IgA FC receptor | | 著者 | ELETSKY, A, CHEN, C, FONG, J.J, NIZET, V, VARKI, A, PRESTEGARD, J.H. | | 登録日 | 2017-06-26 | | 公開日 | 2018-06-27 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | SOLUTION NMR STRUCTURE OF SIGLEC-5 BINDING DOMAIN FROM STREPTOCOCCAL BETA PROTEIN
To Be Published
|
|
7F8S
 
 | | Pennisetum glaucum (Pearl millet) dehydroascorbate reductase (DHAR) with catalytic cysteine (Cy20) in sulphenic and sulfinic acid forms. | | 分子名称: | Dehydroascorbate reductase, SULFATE ION | | 著者 | Das, B.K, Kumar, A, Sreeshma, N.S, Arockiasamy, A. | | 登録日 | 2021-07-02 | | 公開日 | 2022-01-19 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.63 Å) | | 主引用文献 | Comparative kinetic analysis of ascorbate (Vitamin-C) recycling dehydroascorbate reductases from plants and humans.
Biochem.Biophys.Res.Commun., 591, 2021
|
|
5Z79
 
 | | Crystal Structure Analysis of the HPPK-DHPS in complex with substrates | | 分子名称: | 4-AMINOBENZOIC ACID, 5'-DEOXYADENOSINE, 6-HYDROXYMETHYLPTERIN-DIPHOSPHATE, ... | | 著者 | Manickam, Y, Karl, H, Sharma, A. | | 登録日 | 2018-01-27 | | 公開日 | 2018-08-29 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Structure of 6-hydroxymethyl-7,8-dihydropterin pyrophosphokinase-dihydropteroate synthase fromPlasmodium vivaxsheds light on drug resistance
J. Biol. Chem., 293, 2018
|
|
7EVU
 
 | | Co-crystal Structure of Toxoplasma gondii Prolyl tRNA Synthetase (TgPRS) in complex with HFG and JE6 | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 7-bromo-6-chloro-3-{3-[(2R,3S)-3-hydroxypiperidin-2-yl]-2-oxopropyl}quinazolin-4(3H)-one, Prolyl-tRNA synthetase (ProRS), ... | | 著者 | Mishra, S, Malhotra, N, Manickam, Y, Sharma, A. | | 登録日 | 2021-05-22 | | 公開日 | 2022-03-09 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.697 Å) | | 主引用文献 | Double drugging of prolyl-tRNA synthetase provides a new paradigm for anti-infective drug development.
Plos Pathog., 18, 2022
|
|
5D8M
 
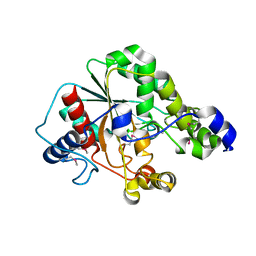 | | Crystal structure of the metagenomic carboxyl esterase MGS0156 | | 分子名称: | Metagenomic carboxyl esterase MGS0156 | | 著者 | Cui, H, Nocek, B, Tchigvintsev, A, Popovic, A, Savchenko, A, Joachimiak, A, Yakunin, A. | | 登録日 | 2015-08-17 | | 公開日 | 2016-10-05 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Crystal structure of esterase (MGS0156)
To Be Published
|
|
6O3K
 
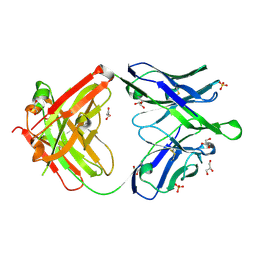 | |
6O3U
 
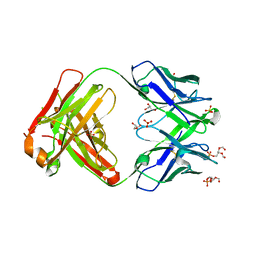 | |
6YXB
 
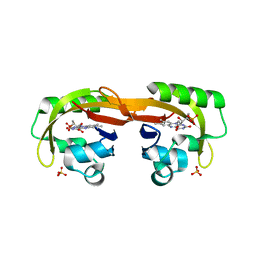 | | Structure of Chloroflexus aggregans flavin based fluorescent protein (CagFbFP) Q148K variant (space group P21) | | 分子名称: | FLAVIN MONONUCLEOTIDE, Multi-sensor hybrid histidine kinase, SULFATE ION | | 著者 | Remeeva, A, Nazarenko, V, Kovalev, K, Gushchin, I. | | 登録日 | 2020-04-30 | | 公開日 | 2021-04-21 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | The molecular basis of spectral tuning in blue- and red-shifted flavin-binding fluorescent proteins.
J.Biol.Chem., 296, 2021
|
|
6YX6
 
 | | Structure of Chloroflexus aggregans flavin based fluorescent protein (CagFbFP) Q148K variant (no morpholine) | | 分子名称: | FLAVIN MONONUCLEOTIDE, Multi-sensor hybrid histidine kinase, SULFATE ION | | 著者 | Remeeva, A, Nazarenko, V, Kovalev, K, Gushchin, I. | | 登録日 | 2020-04-30 | | 公開日 | 2021-04-21 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | The molecular basis of spectral tuning in blue- and red-shifted flavin-binding fluorescent proteins.
J.Biol.Chem., 296, 2021
|
|
6YX4
 
 | | Structure of Chloroflexus aggregans flavin based fluorescent protein (CagFbFP) Q148K variant with morpholine | | 分子名称: | FLAVIN MONONUCLEOTIDE, GLYCEROL, Multi-sensor hybrid histidine kinase, ... | | 著者 | Remeeva, A, Nazarenko, V, Kovalev, K, Gushchin, I. | | 登録日 | 2020-04-30 | | 公開日 | 2021-04-21 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.36 Å) | | 主引用文献 | The molecular basis of spectral tuning in blue- and red-shifted flavin-binding fluorescent proteins.
J.Biol.Chem., 296, 2021
|
|
6O3D
 
 | |
6O3J
 
 | | Crystal structure of the Fab fragment of the human HIV-1 neutralizing antibody PGZL1 in complex with its MPER peptide epitope (region 671-683 of HIV-1 gp41) and phosphatidic acid (06:0 PA) | | 分子名称: | (2R)-3-(phosphonooxy)propane-1,2-diyl dihexanoate, (4S)-2-METHYL-2,4-PENTANEDIOL, MPER peptide, ... | | 著者 | Irimia, A, Wilson, I.A. | | 登録日 | 2019-02-26 | | 公開日 | 2019-12-04 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (3.416 Å) | | 主引用文献 | An MPER antibody neutralizes HIV-1 using germline features shared among donors.
Nat Commun, 10, 2019
|
|
