8WLQ
 
 | | Cryo-EM structure of the whole rod-export apparatus with hook within the flagellar motor-hook complex in the CCW state. | | 分子名称: | Flagellar M-ring protein, Flagellar basal body rod protein FlgB, Flagellar basal-body rod protein FlgC, ... | | 著者 | Tan, J.X, Zhang, L, Zhou, Y, Zhu, Y.Q. | | 登録日 | 2023-09-30 | | 公開日 | 2024-09-04 | | 最終更新日 | 2025-03-26 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Structural basis of the bacterial flagellar motor rotational switching.
Cell Res., 34, 2024
|
|
8WO5
 
 | | Cryo-EM structure of the intact flagellar motor-hook complex in the CCW state | | 分子名称: | Flagellar L-ring protein, Flagellar M-ring protein, Flagellar P-ring protein, ... | | 著者 | Tan, J.X, Zhang, L, Zhou, Y, Zhu, Y.Q. | | 登録日 | 2023-10-06 | | 公開日 | 2024-09-04 | | 最終更新日 | 2025-03-26 | | 実験手法 | ELECTRON MICROSCOPY (7.4 Å) | | 主引用文献 | Structural basis of the bacterial flagellar motor rotational switching.
Cell Res., 34, 2024
|
|
8WLT
 
 | | Cryo-EM structure of the membrane-anchored part of the flagellar motor-hook complex in the CCW state | | 分子名称: | Flagellar L-ring protein, Flagellar M-ring protein, Flagellar P-ring protein, ... | | 著者 | Tan, J.X, Zhang, L, Zhou, Y, Zhu, Y.Q. | | 登録日 | 2023-10-01 | | 公開日 | 2024-09-04 | | 最終更新日 | 2025-03-26 | | 実験手法 | ELECTRON MICROSCOPY (4.1 Å) | | 主引用文献 | Structural basis of the bacterial flagellar motor rotational switching.
Cell Res., 34, 2024
|
|
8WOE
 
 | | Cryo-EM structure of the intact flagellar motor-hook complex in the CW state | | 分子名称: | Chemotaxis protein CheY, Flagellar L-ring protein, Flagellar M-ring protein, ... | | 著者 | Tan, J.X, Zhang, L, Zhou, Y, Zhu, Y.Q. | | 登録日 | 2023-10-07 | | 公開日 | 2024-09-04 | | 最終更新日 | 2025-03-26 | | 実験手法 | ELECTRON MICROSCOPY (4.3 Å) | | 主引用文献 | Structural basis of the bacterial flagellar motor rotational switching.
Cell Res., 34, 2024
|
|
8WLP
 
 | | Cryo-EM structure of the distal rod-hook within the flagellar motor-hook complex in the CCW state. | | 分子名称: | Flagellar basal-body rod protein FlgG, Flagellar hook protein FlgE | | 著者 | Tan, J.X, Zhang, L, Zhou, Y, Zhu, Y.Q. | | 登録日 | 2023-09-30 | | 公開日 | 2024-09-04 | | 最終更新日 | 2025-03-26 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Structural basis of the bacterial flagellar motor rotational switching.
Cell Res., 34, 2024
|
|
8WKK
 
 | | Cryo-EM structure of the whole rod with export apparatus and hook within the flagellar motor-hook complex in the CW state. | | 分子名称: | Flagellar M-ring protein, Flagellar basal body rod protein FlgB, Flagellar basal-body rod protein FlgC, ... | | 著者 | Tan, J.X, Zhang, L, Zhou, Y, Zhu, Y.Q. | | 登録日 | 2023-09-28 | | 公開日 | 2024-09-04 | | 最終更新日 | 2025-03-26 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Structural basis of the bacterial flagellar motor rotational switching.
Cell Res., 34, 2024
|
|
8WK4
 
 | | Cryo-EM structure of the MS ring with FlgB and FliE within the flagellar motor-hook complex in the CW state. | | 分子名称: | Flagellar M-ring protein, Flagellar basal body rod protein FlgB, Flagellar hook-basal body complex protein FliE | | 著者 | Tan, J.X, Zhang, L, Zhou, Y, Zhu, Y.Q. | | 登録日 | 2023-09-27 | | 公開日 | 2024-09-04 | | 最終更新日 | 2025-03-26 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Structural basis of the bacterial flagellar motor rotational switching.
Cell Res., 34, 2024
|
|
8WLN
 
 | | Cryo-EM structure of the MS ring with export apparatus and proximal rod within the motor-hook complex in the CCW state | | 分子名称: | Flagellar M-ring protein, Flagellar basal body rod protein FlgB, Flagellar basal-body rod protein FlgC, ... | | 著者 | Tan, J.X, Zhang, L, Zhou, Y, Zhu, Y.Q. | | 登録日 | 2023-09-30 | | 公開日 | 2024-09-04 | | 最終更新日 | 2025-03-26 | | 実験手法 | ELECTRON MICROSCOPY (4.3 Å) | | 主引用文献 | Structural basis of the bacterial flagellar motor rotational switching.
Cell Res., 34, 2024
|
|
8WIW
 
 | | Cryo-EM structure of the flagellar C ring in the CW state | | 分子名称: | Chemotaxis protein CheY, Flagellar M-ring protein, Flagellar motor switch protein FliG, ... | | 著者 | Tan, J.X, Zhang, L, Zhou, Y, Zhu, Y.Q. | | 登録日 | 2023-09-25 | | 公開日 | 2024-09-04 | | 最終更新日 | 2025-03-26 | | 実験手法 | ELECTRON MICROSCOPY (5.6 Å) | | 主引用文献 | Structural basis of the bacterial flagellar motor rotational switching.
Cell Res., 34, 2024
|
|
8WKI
 
 | | Cryo-EM structure of the distal rod-hook within the flagellar motor-hook complex in the CW state. | | 分子名称: | Flagellar basal-body rod protein FlgG, Flagellar hook protein FlgE | | 著者 | Tan, J.X, Zhang, L, Zhou, Y, Zhu, Y.Q. | | 登録日 | 2023-09-27 | | 公開日 | 2024-09-04 | | 最終更新日 | 2025-03-26 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Structural basis of the bacterial flagellar motor rotational switching.
Cell Res., 34, 2024
|
|
7C4U
 
 | | MicroED structure of orthorhombic Vancomycin at 1.2 A resolution | | 分子名称: | CHLORIDE ION, Vancomycin, vancosamine-(1-2)-beta-D-glucopyranose | | 著者 | Fan, Q, Zhou, H, Li, X, Wang, J. | | 登録日 | 2020-05-18 | | 公開日 | 2020-08-12 | | 最終更新日 | 2025-04-09 | | 実験手法 | ELECTRON CRYSTALLOGRAPHY (1.2 Å) | | 主引用文献 | Precise Control Over Kinetics of Molecular Assembly: Production of Particles with Tunable Sizes and Crystalline Forms.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
7C4V
 
 | | MicroED structure of anorthic Vancomycin at 1.05 A resolution | | 分子名称: | CHLORIDE ION, Vancomycin, vancosamine-(1-2)-beta-D-glucopyranose | | 著者 | Fan, Q, Zhou, H, Li, X, Wang, J. | | 登録日 | 2020-05-18 | | 公開日 | 2020-08-12 | | 最終更新日 | 2023-11-29 | | 実験手法 | ELECTRON CRYSTALLOGRAPHY (1.05 Å) | | 主引用文献 | Precise Control Over Kinetics of Molecular Assembly: Production of Particles with Tunable Sizes and Crystalline Forms.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
7YUJ
 
 | | Crystal structure of HOIL-1L(365-510) | | 分子名称: | DI(HYDROXYETHYL)ETHER, RanBP-type and C3HC4-type zinc finger-containing protein 1, ZINC ION | | 著者 | Xiao, L, Pan, L. | | 登録日 | 2022-08-17 | | 公開日 | 2023-08-30 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.865 Å) | | 主引用文献 | Mechanistic insights into the enzymatic activity of E3 ligase HOIL-1L and its regulation by the linear ubiquitin chain binding.
Sci Adv, 9, 2023
|
|
7YUI
 
 | |
7DWB
 
 | | Human Pannexin1 model | | 分子名称: | Pannexin-1 | | 著者 | Zhang, S.S, Yang, M.J. | | 登録日 | 2021-01-17 | | 公開日 | 2021-05-26 | | 最終更新日 | 2024-10-23 | | 実験手法 | ELECTRON MICROSCOPY (3.15 Å) | | 主引用文献 | Structure of the full-length human Pannexin1 channel and insights into its role in pyroptosis.
Cell Discov, 7, 2021
|
|
8YJT
 
 | | Cryo-EM structure of the flagellar C ring in the CCW state | | 分子名称: | Flagellar M-ring protein, Flagellar motor switch protein FliG, Flagellar motor switch protein FliM, ... | | 著者 | Tan, J.X, Zhang, L, Zhou, Y, Zhu, Y.Q. | | 登録日 | 2024-03-02 | | 公開日 | 2024-09-04 | | 最終更新日 | 2025-03-26 | | 実験手法 | ELECTRON MICROSCOPY (5.9 Å) | | 主引用文献 | Structural basis of the bacterial flagellar motor rotational switching.
Cell Res., 34, 2024
|
|
8XP1
 
 | | Cryo-EM structure of the protomers of the C ring in the CW state | | 分子名称: | Chemotaxis protein CheY, Flagellar M-ring protein, Flagellar motor switch protein FliG, ... | | 著者 | Tan, J.X, Zhang, L, Zhou, Y, Zhu, Y.Q. | | 登録日 | 2024-01-02 | | 公開日 | 2024-09-04 | | 最終更新日 | 2025-03-26 | | 実験手法 | ELECTRON MICROSCOPY (4.4 Å) | | 主引用文献 | Structural basis of the bacterial flagellar motor rotational switching.
Cell Res., 34, 2024
|
|
8XP0
 
 | | Cryo-EM structure of the protomers of the C ring in the CCW state | | 分子名称: | Flagellar M-ring protein, Flagellar motor switch protein FliG, Flagellar motor switch protein FliM, ... | | 著者 | Tan, J.X, Zhang, L, Zhou, Y, Zhu, Y.Q. | | 登録日 | 2024-01-02 | | 公開日 | 2024-09-04 | | 最終更新日 | 2025-03-26 | | 実験手法 | ELECTRON MICROSCOPY (4 Å) | | 主引用文献 | Structural basis of the bacterial flagellar motor rotational switching.
Cell Res., 34, 2024
|
|
5YGI
 
 | |
2JV0
 
 | |
7E1V
 
 | | Cryo-EM structure of apo hybrid respiratory supercomplex consisting of Mycobacterium tuberculosis complexIII and Mycobacterium smegmatis complexIV | | 分子名称: | (2R)-2-(hexadecanoyloxy)-3-{[(S)-hydroxy{[(1R,2R,3R,4R,5R,6S)-2,3,4,5,6-pentahydroxycyclohexyl]oxy}phosphoryl]oxy}propyl (9S)-9-methyloctadecanoate, (2R)-3-(((2-aminoethoxy)(hydroxy)phosphoryl)oxy)-2-(palmitoyloxy)propyl (E)-octadec-9-enoate, CARDIOLIPIN, ... | | 著者 | Zhou, S, Wang, W, Gao, Y, Gong, H, Rao, Z. | | 登録日 | 2021-02-03 | | 公開日 | 2021-10-13 | | 最終更新日 | 2025-07-02 | | 実験手法 | ELECTRON MICROSCOPY (2.68 Å) | | 主引用文献 | Structure of Mycobacterium tuberculosis cytochrome bcc in complex with Q203 and TB47, two anti-TB drug candidates.
Elife, 10, 2021
|
|
7E1X
 
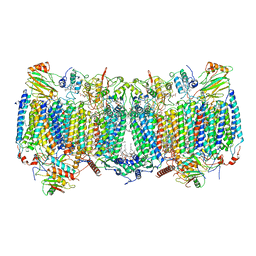 | | Cryo-EM structure of hybrid respiratory supercomplex consisting of Mycobacterium tuberculosis complexIII and Mycobacterium smegmatis complexIV in presence of TB47 | | 分子名称: | (2R)-2-(hexadecanoyloxy)-3-{[(S)-hydroxy{[(1R,2R,3R,4R,5R,6S)-2,3,4,5,6-pentahydroxycyclohexyl]oxy}phosphoryl]oxy}propyl (9S)-9-methyloctadecanoate, (2R)-3-(((2-aminoethoxy)(hydroxy)phosphoryl)oxy)-2-(palmitoyloxy)propyl (E)-octadec-9-enoate, 5-methoxy-2-methyl-~{N}-[[4-[4-[4-(trifluoromethyloxy)phenyl]piperidin-1-yl]phenyl]methyl]pyrazolo[1,5-a]pyridine-3-carboxamide, ... | | 著者 | Zhou, S, Wang, W, Gao, Y, Gong, H, Rao, Z. | | 登録日 | 2021-02-03 | | 公開日 | 2021-10-27 | | 最終更新日 | 2024-11-20 | | 実験手法 | ELECTRON MICROSCOPY (2.93 Å) | | 主引用文献 | Structure of Mycobacterium tuberculosis cytochrome bcc in complex with Q203 and TB47, two anti-TB drug candidates.
Elife, 10, 2021
|
|
7E1W
 
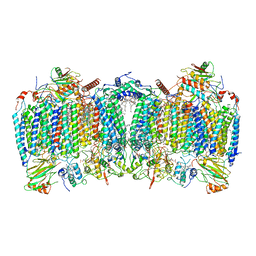 | | Cryo-EM structure of hybrid respiratory supercomplex consisting of Mycobacterium tuberculosis complexIII and Mycobacterium smegmatis complexIV in the presence of Q203 | | 分子名称: | (2R)-2-(hexadecanoyloxy)-3-{[(S)-hydroxy{[(1R,2R,3R,4R,5R,6S)-2,3,4,5,6-pentahydroxycyclohexyl]oxy}phosphoryl]oxy}propyl (9S)-9-methyloctadecanoate, (2R)-3-(((2-aminoethoxy)(hydroxy)phosphoryl)oxy)-2-(palmitoyloxy)propyl (E)-octadec-9-enoate, 6-chloranyl-2-ethyl-N-[[4-[4-[4-(trifluoromethyloxy)phenyl]piperidin-1-yl]phenyl]methyl]imidazo[1,2-a]pyridine-3-carboxamide, ... | | 著者 | Zhou, S, Wang, W, Gao, Y, Gong, H, Rao, Z. | | 登録日 | 2021-02-03 | | 公開日 | 2021-10-27 | | 最終更新日 | 2024-10-16 | | 実験手法 | ELECTRON MICROSCOPY (2.67 Å) | | 主引用文献 | Structure of Mycobacterium tuberculosis cytochrome bcc in complex with Q203 and TB47, two anti-TB drug candidates.
Elife, 10, 2021
|
|
2MN4
 
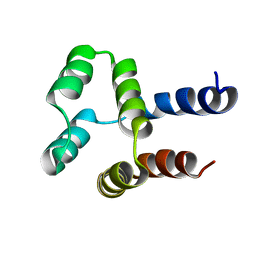 | | NMR solution structure of a computational designed protein based on structure template 1cy5 | | 分子名称: | Computational designed protein based on structure template 1cy5 | | 著者 | Xiong, P, Wang, M, Zhang, J, Chen, Q, Liu, H. | | 登録日 | 2014-03-28 | | 公開日 | 2014-10-29 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Protein design with a comprehensive statistical energy function and boosted by experimental selection for foldability
Nat Commun, 5, 2014
|
|
2MLB
 
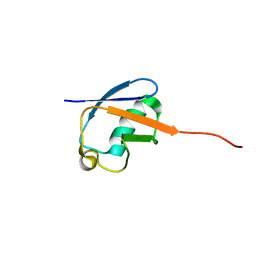 | | NMR solution structure of a computational designed protein based on template of human erythrocytic ubiquitin | | 分子名称: | redesigned ubiquitin | | 著者 | Xiong, P, Wang, M, Zhang, J, Chen, Q, Liu, H. | | 登録日 | 2014-02-21 | | 公開日 | 2014-10-29 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Protein design with a comprehensive statistical energy function and boosted by experimental selection for foldability
Nat Commun, 5, 2014
|
|
