3WB8
 
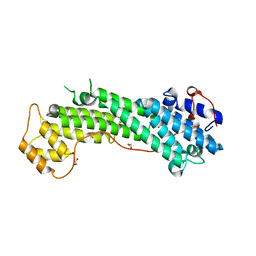 | | Crystal Structure of MyoVa-GTD | | 分子名称: | 1,2-ETHANEDIOL, Unconventional myosin-Va | | 著者 | Wei, Z, Liu, X, Yu, C, Zhang, M. | | 登録日 | 2013-05-13 | | 公開日 | 2013-07-10 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.499 Å) | | 主引用文献 | Structural basis of cargo recognitions for class V myosins
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3CXS
 
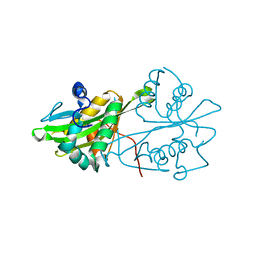 | | Crystal structure of human GNA1 | | 分子名称: | Glucosamine 6-phosphate N-acetyltransferase | | 著者 | Wang, J, Liu, X, Li, L.-F, Su, X.-D. | | 登録日 | 2008-04-25 | | 公開日 | 2008-09-16 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Acceptor substrate binding revealed by crystal structure of human glucosamine-6-phosphate N-acetyltransferase 1
Febs Lett., 582, 2008
|
|
3EXS
 
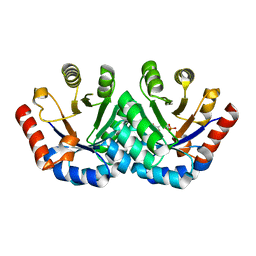 | | Crystal structure of KGPDC from Streptococcus mutans in complex with D-R5P | | 分子名称: | RIBULOSE-5-PHOSPHATE, RmpD (Hexulose-6-phosphate synthase) | | 著者 | Li, G.L, Liu, X, Wang, K.T, Li, L.F, Su, X.D. | | 登録日 | 2008-10-17 | | 公開日 | 2009-08-25 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Open-closed conformational change revealed by the crystal structures of 3-keto-L-gulonate 6-phosphate decarboxylase from Streptococcus mutans
Biochem.Biophys.Res.Commun., 381, 2009
|
|
3CXQ
 
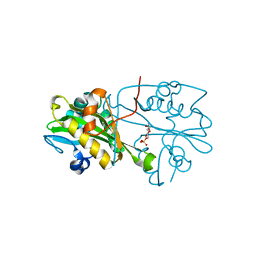 | | Crystal structure of human glucosamine 6-phosphate N-acetyltransferase 1 bound to GlcN6P | | 分子名称: | 2-amino-2-deoxy-6-O-phosphono-alpha-D-glucopyranose, Glucosamine 6-phosphate N-acetyltransferase | | 著者 | Wang, J, Liu, X, Li, L.-F, Su, X.-D. | | 登録日 | 2008-04-25 | | 公開日 | 2008-09-16 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Acceptor substrate binding revealed by crystal structure of human glucosamine-6-phosphate N-acetyltransferase 1
Febs Lett., 582, 2008
|
|
6JKM
 
 | | Crystal structure of BubR1 kinase domain | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, MAGNESIUM ION, ... | | 著者 | Lin, L, Ye, S, Huang, Y, Liu, X, Zhang, R, Yao, X. | | 登録日 | 2019-03-01 | | 公開日 | 2019-06-26 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | BubR1 phosphorylates CENP-E as a switch enabling the transition from lateral association to end-on capture of spindle microtubules.
Cell Res., 29, 2019
|
|
6VVO
 
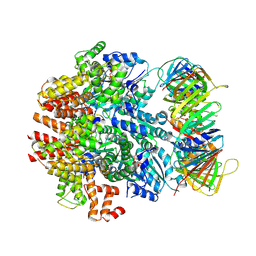 | | Structure of the human clamp loader (Replication Factor C, RFC) bound to the sliding clamp (Proliferating Cell Nuclear Antigen, PCNA) | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | 著者 | Gaubitz, C, Liu, X, Stone, N.P, Kelch, B.A. | | 登録日 | 2020-02-18 | | 公開日 | 2020-02-26 | | 最終更新日 | 2020-03-25 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Structure of the human clamp loader bound to the sliding clamp: a further twist on AAA+ mechanism
Biorxiv, 2020
|
|
6JKK
 
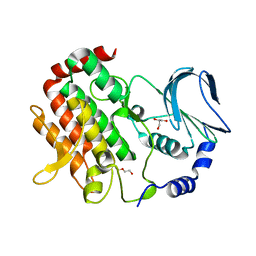 | | Crystal structure of BubR1 kinase domain | | 分子名称: | DI(HYDROXYETHYL)ETHER, GLYCEROL, Mitotic checkpoint control protein kinase BUB1 | | 著者 | Lin, L, Ye, S, Huang, Y, Liu, X, Zhang, R, Yao, X. | | 登録日 | 2019-03-01 | | 公開日 | 2019-06-26 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | BubR1 phosphorylates CENP-E as a switch enabling the transition from lateral association to end-on capture of spindle microtubules.
Cell Res., 29, 2019
|
|
4ANJ
 
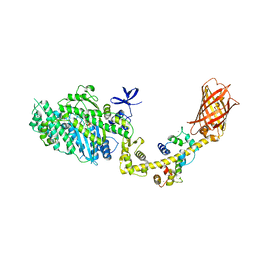 | | MYOSIN VI (MDinsert2-GFP fusion) PRE-POWERSTROKE STATE (MG.ADP.AlF4) | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, CALCIUM ION, CALMODULIN, ... | | 著者 | Menetrey, J, Isabet, T, Ropars, V, Mukherjea, M, Pylypenko, O, Liu, X, Perez, J, Vachette, P, Sweeney, H.L, Houdusse, A.M. | | 登録日 | 2012-03-19 | | 公開日 | 2012-10-17 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Processive Steps in the Reverse Direction Require Uncoupling of the Lead Head Lever Arm of Myosin Vi.
Mol.Cell, 48, 2012
|
|
3V69
 
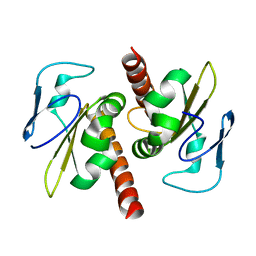 | | Filia-N crystal structure | | 分子名称: | Protein Filia | | 著者 | Wang, J, Xu, M, Zhu, K, Li, L, Liu, X. | | 登録日 | 2011-12-19 | | 公開日 | 2012-02-15 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | The N-terminus of FILIA Forms an Atypical KH Domain with a Unique Extension Involved in Interaction with RNA.
Plos One, 7, 2012
|
|
4LSW
 
 | |
7TBI
 
 | | Composite structure of the S. cerevisiae nuclear pore complex (NPC) | | 分子名称: | Dyn2, Nic96 R1, Nic96 R2, ... | | 著者 | Petrovic, S, Samanta, D, Perriches, T, Bley, C.J, Thierbach, K, Brown, B, Nie, S, Mobbs, G.W, Stevens, T.A, Liu, X, Tomaleri, G.P, Schaus, L, Hoelz, A. | | 登録日 | 2021-12-22 | | 公開日 | 2022-06-15 | | 最終更新日 | 2022-06-22 | | 実験手法 | ELECTRON MICROSCOPY (25 Å) | | 主引用文献 | Architecture of the linker-scaffold in the nuclear pore.
Science, 376, 2022
|
|
4ROU
 
 | |
5JPU
 
 | | Structure of limonene epoxide hydrolase mutant - H-2-H5 complex with (S,S)-cyclohexane-1,2-diol | | 分子名称: | (1S,2S)-cyclohexane-1,2-diol, limonene epoxide hydrolase | | 著者 | Li, G, Zhang, H, Sun, Z, Liu, X, Reetz, M.T. | | 登録日 | 2016-05-04 | | 公開日 | 2016-06-15 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Multi-Parameter Optimization in Directed Evolution: Engineering Thermostability, Enantioselectivity and Activity of an Epoxide Hydrolase
To be published
|
|
4QQR
 
 | |
4D3E
 
 | | Tetramer of IpaD, modified from 2J0O, fitted into negative stain electron microscopy reconstruction of the wild type tip complex from the type III secretion system of Shigella flexneri | | 分子名称: | INVASIN IPAD | | 著者 | Cheung, M, Shen, D.-K, Makino, F, Kato, T, Roehrich, D, Martinez-Argudo, I, Walker, M.L, Murillo, I, Liu, X, Pain, M, Brown, J, Frazer, G, Mantell, J, Mina, P, Todd, T, Sessions, R.B, Namba, K, Blocker, A.J. | | 登録日 | 2014-10-21 | | 公開日 | 2014-12-10 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (24 Å) | | 主引用文献 | Three-Dimensional Electron Microscopy Reconstruction and Cysteine-Mediated Crosslinking Provide a Model of the T3Ss Needle Tip Complex.
Mol.Microbiol., 95, 2015
|
|
3L7W
 
 | |
3L9T
 
 | |
3LAS
 
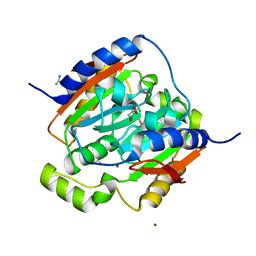 | | Crystal structure of carbonic anhydrase from streptococcus mutans to 1.4 angstrom resolution | | 分子名称: | GLYCEROL, GUANIDINE, MAGNESIUM ION, ... | | 著者 | Ma, L.-L, Wang, K.-T, Liu, X, Su, X.-D. | | 登録日 | 2010-01-07 | | 公開日 | 2011-01-12 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Crystal structure of carbonic anhydrase from streptococcus mutans to 1.4 angstrom resolution
To be published
|
|
3L7X
 
 | |
3L87
 
 | |
3L9C
 
 | |
3LA8
 
 | |
3LBB
 
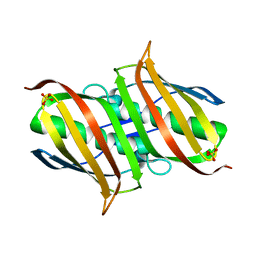 | | The Crystal Structure of smu.793 from Streptococcus mutans UA159 | | 分子名称: | CHLORIDE ION, Putative uncharacterized protein smu.793, SULFATE ION | | 著者 | Su, X.-D, Hou, Q.M, Fan, X.X, Nan, J, Liu, X. | | 登録日 | 2010-01-08 | | 公開日 | 2011-01-12 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | The Crystal Structure of smu.793 from Streptococcus mutans UA159
TO BE PUBLISHED
|
|
3LBA
 
 | |
6M49
 
 | | cryo-EM structure of Scap/Insig complex in the present of 25-hydroxyl cholesterol. | | 分子名称: | 25-HYDROXYCHOLESTEROL, Insulin-induced gene 2 protein, Sterol regulatory element-binding protein cleavage-activating protein,Sterol regulatory element-binding protein cleavage-activating protein | | 著者 | Yan, R, Cao, P, Song, W, Qian, H, Du, X, Coates, H.W, Zhao, X, Li, Y, Gao, S, Gong, X, Liu, X, Sui, J, Lei, J, Yang, H, Brown, A.J, Zhou, Q, Yan, C, Yan, N. | | 登録日 | 2020-03-06 | | 公開日 | 2021-01-20 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | A structure of human Scap bound to Insig-2 suggests how their interaction is regulated by sterols.
Science, 371, 2021
|
|
