8YZZ
 
 | | The structure of HLA-A*2402 complex with peptide from SARS-CoV-2 S448-456 NYNYLYRLF(Prototype) | | 分子名称: | Beta-2-microglobulin, MHC class I antigen, Spike protein S1 | | 著者 | Shang, B.L, Zhang, J.N, Tian, J.M, Liu, J. | | 登録日 | 2024-04-08 | | 公開日 | 2025-01-29 | | 最終更新日 | 2025-05-28 | | 実験手法 | X-RAY DIFFRACTION (1.88 Å) | | 主引用文献 | T cell immune evasion by SARS-CoV-2 JN.1 escapees targeting two cytotoxic T cell epitope hotspots.
Nat.Immunol., 26, 2025
|
|
8Z07
 
 | |
8Z06
 
 | |
8Z05
 
 | | The structure of HLA-A*0201 complex with peptide from SARS-CoV-2 N222-230 LLLDRLNKL(BA.2.86/JN.1) | | 分子名称: | Beta-2-microglobulin, HLA class I histocompatibility antigen, A alpha chain, ... | | 著者 | Zhang, J.N, Tian, J.M, Liu, J. | | 登録日 | 2024-04-09 | | 公開日 | 2025-01-29 | | 最終更新日 | 2025-05-28 | | 実験手法 | X-RAY DIFFRACTION (1.96 Å) | | 主引用文献 | T cell immune evasion by SARS-CoV-2 JN.1 escapees targeting two cytotoxic T cell epitope hotspots.
Nat.Immunol., 26, 2025
|
|
8XK2
 
 | | A neutralizing nanobody VHH60 against wt SARS-CoV-2 | | 分子名称: | Spike protein S1, VHH60 nanobody | | 著者 | Lu, Y, Guo, H, Ji, X, Yang, H. | | 登録日 | 2023-12-22 | | 公開日 | 2024-07-10 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (3.4 Å) | | 主引用文献 | A broad neutralizing nanobody against SARS-CoV-2 engineered from an approved drug.
Cell Death Dis, 15, 2024
|
|
8XKI
 
 | | A neutralizing nanobody VHH60 against wt SARS-CoV-2 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | 著者 | Lu, Y, Guo, H, Ji, X, Yang, H. | | 登録日 | 2023-12-23 | | 公開日 | 2024-07-10 | | 最終更新日 | 2025-07-02 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | A broad neutralizing nanobody against SARS-CoV-2 engineered from an approved drug.
Cell Death Dis, 15, 2024
|
|
8Z08
 
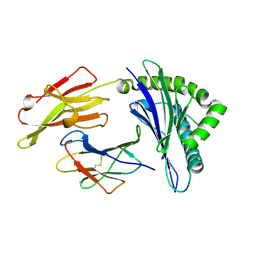 | |
8YZW
 
 | |
7WM0
 
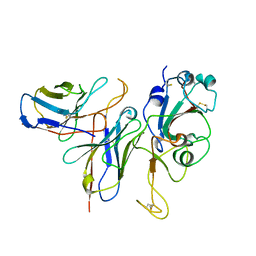 | |
4FLN
 
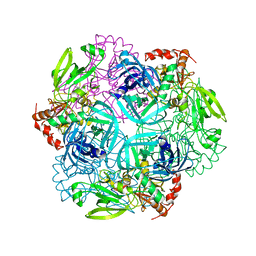 | | Crystal structure of plant protease Deg2 | | 分子名称: | Protease Do-like 2, chloroplastic, Unknown peptide | | 著者 | Gong, W, Liu, L, Sun, R, Gao, F. | | 登録日 | 2012-06-15 | | 公開日 | 2012-09-19 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Crystal structure of Arabidopsis deg2 protein reveals an internal PDZ ligand locking the hexameric resting state.
J.Biol.Chem., 287, 2012
|
|
7WLY
 
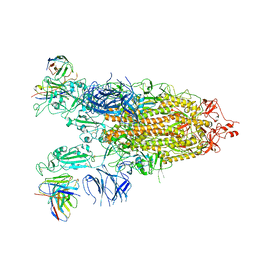 | | Cryo-EM structure of the Omicron S in complex with 35B5 Fab(1 down- and 2 up RBDs) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of 35B5 Fab, ... | | 著者 | Wang, X, Zhu, Y. | | 登録日 | 2022-01-14 | | 公開日 | 2022-05-25 | | 最終更新日 | 2024-11-13 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | 35B5 antibody potently neutralizes SARS-CoV-2 Omicron by disrupting the N-glycan switch via a conserved spike epitope.
Cell Host Microbe, 30, 2022
|
|
7WLZ
 
 | |
2MOG
 
 | | Solution structure of the terminal Ig-like domain from Leptospira interrogans LigB | | 分子名称: | Bacterial Ig-like domain, group 2 | | 著者 | Ptak, C.P, Hsieh, C, Lin, Y, Maltsev, A.S, Raman, R, Sharma, Y, Oswald, R.E, Chang, Y. | | 登録日 | 2014-04-25 | | 公開日 | 2014-08-13 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | NMR Solution Structure of the Terminal Immunoglobulin-like Domain from the Leptospira Host-Interacting Outer Membrane Protein, LigB.
Biochemistry, 53, 2014
|
|
2DHO
 
 | | Crystal structure of human IPP isomerase I in space group P212121 | | 分子名称: | CHLORIDE ION, Isopentenyl-diphosphate delta-isomerase 1, MANGANESE (II) ION, ... | | 著者 | Zhang, C, Wei, Z, Gong, W. | | 登録日 | 2006-03-24 | | 公開日 | 2007-06-05 | | 最終更新日 | 2024-11-20 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Crystal structures of human IPP isomerase: new insights into the catalytic mechanism
J.Mol.Biol., 366, 2007
|
|
2WLA
 
 | | Streptococcus pyogenes Dpr | | 分子名称: | DPS-LIKE PEROXIDE RESISTANCE PROTEIN, GLYCEROL | | 著者 | Haikarainen, T, Tsou, C.-C, Wu, J.-J, Papageorgiou, A.C. | | 登録日 | 2009-06-23 | | 公開日 | 2009-09-15 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystal Structures of Streptococcus Pyogenes Dpr Reveal a Dodecameric Iron-Binding Protein with a Ferroxidase Site.
J.Biol.Inorg.Chem., 15, 2010
|
|
8Z96
 
 | | Crystal structure of CrtAgo/TIR-APAZ in complex with guide DNA and 21-nt target DNA | | 分子名称: | DNA (5'-D(*CP*TP*AP*TP*AP*CP*AP*AP*CP*CP*TP*AP*CP*TP*AP*CP*CP*TP*CP*AP*T)-3'), DNA (5'-D(P*TP*GP*AP*GP*GP*TP*AP*GP*TP*AP*GP*GP*TP*TP*GP*TP*AP*TP*AP*GP*T)-3'), MAGNESIUM ION, ... | | 著者 | Hu, R, Chen, J, Liu, L. | | 登録日 | 2024-04-22 | | 公開日 | 2025-03-12 | | 実験手法 | X-RAY DIFFRACTION (3.36 Å) | | 主引用文献 | Structural basis of ssDNA-guided NADase activation of prokaryotic SPARTA system.
Nucleic Acids Res., 53, 2025
|
|
8Z92
 
 | | Crystal structure of CrtAgo/TIR-APAZ in complex with guide DNA and 16-nt target DNA | | 分子名称: | DNA (5'-D(*CP*AP*AP*CP*CP*TP*AP*CP*TP*AP*CP*CP*TP*CP*AP*T)-3'), DNA (5'-D(P*TP*GP*AP*GP*GP*TP*AP*GP*TP*AP*GP*GP*TP*TP*GP*TP*AP*TP*AP*GP*T)-3'), Piwi domain-containing protein, ... | | 著者 | Hu, R, Chen, J, Liu, L. | | 登録日 | 2024-04-22 | | 公開日 | 2025-03-12 | | 実験手法 | X-RAY DIFFRACTION (3.85 Å) | | 主引用文献 | Structural basis of ssDNA-guided NADase activation of prokaryotic SPARTA system.
Nucleic Acids Res., 53, 2025
|
|
8Z8Y
 
 | |
7T9B
 
 | | ApexGT5 in complex with GT5-d42.16 and RM20A3 Fabs | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Berndsen, Z.T, Ward, A.B. | | 登録日 | 2021-12-18 | | 公開日 | 2022-09-28 | | 最終更新日 | 2024-10-30 | | 実験手法 | ELECTRON MICROSCOPY (3.72 Å) | | 主引用文献 | Priming HIV Envelope V2 Apex-directed broadly neutralizing antibody responses with protein or mRNA immunogens
Immunity, 2022
|
|
7T9A
 
 | | ApexGT2 in complex with GT2-d42.16 and RM20A3 Fabs | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GT2-d42.16 Fab Heavy Chain, ... | | 著者 | Berndsen, Z.T, Ward, A.B. | | 登録日 | 2021-12-18 | | 公開日 | 2022-09-28 | | 最終更新日 | 2024-10-30 | | 実験手法 | ELECTRON MICROSCOPY (3.54 Å) | | 主引用文献 | Priming HIV Envelope V2 Apex-directed broadly neutralizing antibody responses with protein or mRNA immunogens
Immunity, 2022
|
|
2WLU
 
 | | Iron-bound crystal structure of Streptococcus pyogenes Dpr | | 分子名称: | DPS-LIKE PEROXIDE RESISTANCE PROTEIN, FE (III) ION, GLYCEROL, ... | | 著者 | Haikarainen, T, Tsou, C.-C, Wu, J.-J, Papageorgiou, A.C. | | 登録日 | 2009-06-26 | | 公開日 | 2009-09-15 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.94 Å) | | 主引用文献 | Crystal Structures of Streptococcus Pyogenes Dpr Reveal a Dodecameric Iron-Binding Protein with a Ferroxidase Site.
J.Biol.Inorg.Chem., 15, 2010
|
|
8XG2
 
 | | The structure of HLA-A/Pep14 | | 分子名称: | Beta-2-microglobulin, HLA class I heavy chain, Spike protein S1 | | 著者 | Zhang, J.N, Yue, C, Liu, J. | | 登録日 | 2023-12-14 | | 公開日 | 2024-07-10 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (1.84 Å) | | 主引用文献 | Uncommon P1 Anchor-featured Viral T Cell Epitope Preference within HLA-A*2601 and HLA-A*0101 Individuals.
Immunohorizons, 8, 2024
|
|
8XKE
 
 | | The structure of HLA-A/14-3-D | | 分子名称: | Beta-2-microglobulin, GLU-VAL-ASP-ASN-ALA-THR-ARG-PHE-ALA-SER-VAL-TYR, HLA class I heavy chain | | 著者 | Zhang, J.N, Yue, C, Liu, J, Sun, Z.Y. | | 登録日 | 2023-12-23 | | 公開日 | 2024-07-10 | | 最終更新日 | 2024-11-13 | | 実験手法 | X-RAY DIFFRACTION (1.92 Å) | | 主引用文献 | Uncommon P1 Anchor-featured Viral T Cell Epitope Preference within HLA-A*2601 and HLA-A*0101 Individuals.
Immunohorizons, 8, 2024
|
|
8XFZ
 
 | | The structure of HLA-A/L1-2 | | 分子名称: | Beta-2-microglobulin, HLA class I heavy chain, Major capsid protein L1 | | 著者 | Zhang, J.N, Yue, C, Liu, J. | | 登録日 | 2023-12-14 | | 公開日 | 2024-07-10 | | 最終更新日 | 2024-11-13 | | 実験手法 | X-RAY DIFFRACTION (2.32 Å) | | 主引用文献 | Uncommon P1 Anchor-featured Viral T Cell Epitope Preference within HLA-A*2601 and HLA-A*0101 Individuals.
Immunohorizons, 8, 2024
|
|
8XKC
 
 | | The structure of HLA-A/Pep16 | | 分子名称: | Beta-2-microglobulin, HLA class I heavy chain, Spike protein S1 | | 著者 | Zhang, J.N, Yue, C, Liu, J. | | 登録日 | 2023-12-23 | | 公開日 | 2024-07-10 | | 最終更新日 | 2024-11-13 | | 実験手法 | X-RAY DIFFRACTION (2.18 Å) | | 主引用文献 | Uncommon P1 Anchor-featured Viral T Cell Epitope Preference within HLA-A*2601 and HLA-A*0101 Individuals.
Immunohorizons, 8, 2024
|
|
