5C74
 
 | |
2KBE
 
 | |
2KBF
 
 | |
2Q6F
 
 | | Crystal structure of infectious bronchitis virus (IBV) main protease in complex with a Michael acceptor inhibitor N3 | | 分子名称: | Infectious bronchitis virus (IBV) main protease, N-[(5-METHYLISOXAZOL-3-YL)CARBONYL]ALANYL-L-VALYL-N~1~-((1R,2Z)-4-(BENZYLOXY)-4-OXO-1-{[(3R)-2-OXOPYRROLIDIN-3-YL]METHYL}BUT-2-ENYL)-L-LEUCINAMIDE | | 著者 | Xue, X.Y, Yang, H.T, Xue, F, Bartlam, M, Rao, Z.H. | | 登録日 | 2007-06-05 | | 公開日 | 2008-02-12 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structures of two coronavirus main proteases: implications for substrate binding and antiviral drug design.
J.Virol., 82, 2008
|
|
2Q6D
 
 | | Crystal structure of infectious bronchitis virus (IBV) main protease | | 分子名称: | Infectious bronchitis virus (IBV) main protease | | 著者 | Xue, X.Y, Yang, H.T, Xue, F, Bartlam, M, Rao, Z.H. | | 登録日 | 2007-06-04 | | 公開日 | 2008-02-12 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | Structures of two coronavirus main proteases: implications for substrate binding and antiviral drug design.
J.Virol., 82, 2008
|
|
2Q6G
 
 | | Crystal structure of SARS-CoV main protease H41A mutant in complex with an N-terminal substrate | | 分子名称: | Polypeptide chain, severe acute respiratory syndrome coronavirus (SARS-CoV) | | 著者 | Xue, X.Y, Yang, H.T, Xue, F, Bartlam, M, Rao, Z.H. | | 登録日 | 2007-06-05 | | 公開日 | 2008-02-12 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structures of two coronavirus main proteases: implications for substrate binding and antiviral drug design.
J.Virol., 82, 2008
|
|
5X0W
 
 | | Molecular mechanism for the binding between Sharpin and HOIP | | 分子名称: | E3 ubiquitin-protein ligase RNF31, Sharpin | | 著者 | Liu, J, Li, F, Cheng, X, Pan, L. | | 登録日 | 2017-01-23 | | 公開日 | 2017-10-18 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Structural Insights into SHARPIN-Mediated Activation of HOIP for the Linear Ubiquitin Chain Assembly
Cell Rep, 21, 2017
|
|
4YUS
 
 | | Crystal structure of photoactivated adenylyl cyclase of a cyanobacteriaOscillatoria acuminata in hexagonal form | | 分子名称: | FLAVIN MONONUCLEOTIDE, Family 3 adenylate cyclase | | 著者 | Park, S.-Y, Ohki, M, Sugiyama, K, Kawai, F, Iseki, M. | | 登録日 | 2015-03-19 | | 公開日 | 2016-06-01 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structural insight into photoactivation of an adenylate cyclase from a photosynthetic cyanobacterium
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5YDR
 
 | | Structure of DNMT1 RFTS domain in complex with ubiquitin | | 分子名称: | DNA (cytosine-5)-methyltransferase 1, PHOSPHATE ION, Polyubiquitin-B, ... | | 著者 | Qian, C. | | 登録日 | 2017-09-14 | | 公開日 | 2018-02-21 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.003 Å) | | 主引用文献 | Structural and mechanistic insights into UHRF1-mediated DNMT1 activation in the maintenance DNA methylation.
Nucleic Acids Res., 46, 2018
|
|
4YUT
 
 | | Crystal structure of photoactivated adenylyl cyclase of a cyanobacteriaOscillatoria acuminata in orthorhombic form | | 分子名称: | FLAVIN MONONUCLEOTIDE, Family 3 adenylate cyclase | | 著者 | Park, S.-Y, Ohki, M, Sugiyama, K, Kawai, F, Iseki, M. | | 登録日 | 2015-03-19 | | 公開日 | 2016-06-01 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Structural insight into photoactivation of an adenylate cyclase from a photosynthetic cyanobacterium
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
4Z69
 
 | |
5WQ4
 
 | |
2KLZ
 
 | |
1OXP
 
 | | ASPARTATE AMINOTRANSFERASE, H-ASP COMPLEX, CLOSED CONFORMATION | | 分子名称: | 4'-DEOXY-4'-ACETYLYAMINO-PYRIDOXAL-5'-PHOSPHATE, ASPARTATE AMINOTRANSFERASE | | 著者 | Hohenester, E, Schirmer, T, Jansonius, J.N. | | 登録日 | 1995-12-23 | | 公開日 | 1996-06-10 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Crystal structures and solution studies of oxime adducts of mitochondrial aspartate aminotransferase.
Eur.J.Biochem., 236, 1996
|
|
1OXO
 
 | | ASPARTATE AMINOTRANSFERASE, H-ASP COMPLEX, OPEN CONFORMATION | | 分子名称: | 4'-DEOXY-4'-ACETYLYAMINO-PYRIDOXAL-5'-PHOSPHATE, ASPARTATE AMINOTRANSFERASE | | 著者 | Hohenester, E, Schirmer, T, Jansonius, J.N. | | 登録日 | 1995-12-23 | | 公開日 | 1996-06-10 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Crystal structures and solution studies of oxime adducts of mitochondrial aspartate aminotransferase.
Eur.J.Biochem., 236, 1996
|
|
1PDO
 
 | |
7EP2
 
 | |
7EP0
 
 | | Crystal structure of ZYG11B bound to GSTE degron | | 分子名称: | Protein zyg-11 homolog B, sodium 3,3'-(1E,1'E)-biphenyl-4,4'-diylbis(diazene-2,1-diyl)bis(4-aminonaphthalene-1-sulfonate) | | 著者 | Yan, X, Li, Y. | | 登録日 | 2021-04-26 | | 公開日 | 2021-07-14 | | 最終更新日 | 2021-09-01 | | 実験手法 | X-RAY DIFFRACTION (2.16 Å) | | 主引用文献 | Molecular basis for recognition of Gly/N-degrons by CRL2 ZYG11B and CRL2 ZER1 .
Mol.Cell, 81, 2021
|
|
7EP3
 
 | |
7EP5
 
 | |
7EP1
 
 | |
7EP4
 
 | |
6K08
 
 | |
6K07
 
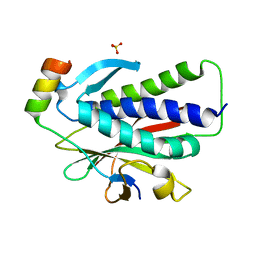 | | Crystal structure of REV7(R124A) in complex with a Shieldin3 fragment | | 分子名称: | Mitotic spindle assembly checkpoint protein MAD2B, SULFATE ION, Shieldin complex subunit 3 | | 著者 | Zhang, F, Dai, Y. | | 登録日 | 2019-05-05 | | 公開日 | 2019-12-11 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.24 Å) | | 主引用文献 | Structural basis for shieldin complex subunit 3-mediated recruitment of the checkpoint protein REV7 during DNA double-strand break repair.
J.Biol.Chem., 295, 2020
|
|
2LJ1
 
 | |
