7VGZ
 
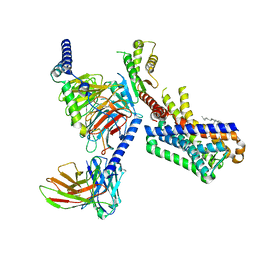 | | MT1-remalteon-Gi complex | | 分子名称: | CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | 著者 | Wang, Q.G, Lu, Q.Y. | | 登録日 | 2021-09-20 | | 公開日 | 2022-03-02 | | 最終更新日 | 2022-09-14 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Structural basis of the ligand binding and signaling mechanism of melatonin receptors.
Nat Commun, 13, 2022
|
|
7VY5
 
 | | Coxsackievirus B3 (VP3-234Q) incubation with CD55 at pH7.4 | | 分子名称: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3, ... | | 著者 | Wang, Q.L, Liu, C.C. | | 登録日 | 2021-11-13 | | 公開日 | 2022-01-19 | | 最終更新日 | 2022-08-10 | | 実験手法 | ELECTRON MICROSCOPY (3.15 Å) | | 主引用文献 | Molecular basis of differential receptor usage for naturally occurring CD55-binding and -nonbinding coxsackievirus B3 strains.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7VXZ
 
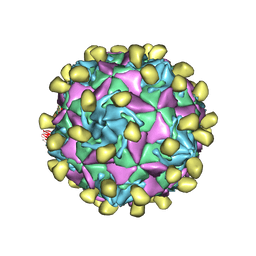 | |
7VYK
 
 | |
7VYL
 
 | |
7VXH
 
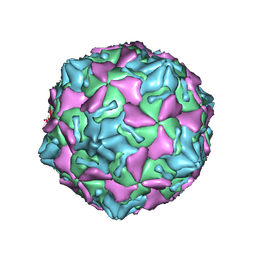 | | Coxsackievirus B3 full particle at pH7.4 (VP3-234Q) | | 分子名称: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3, ... | | 著者 | Wang, Q.L, Liu, C.C. | | 登録日 | 2021-11-12 | | 公開日 | 2022-01-19 | | 最終更新日 | 2024-06-26 | | 実験手法 | ELECTRON MICROSCOPY (2.95 Å) | | 主引用文献 | Molecular basis of differential receptor usage for naturally occurring CD55-binding and -nonbinding coxsackievirus B3 strains.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7VYM
 
 | |
7VY0
 
 | | Coxsackievirus B3 full particle at pH7.4 (VP3-234N) | | 分子名称: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3, ... | | 著者 | Wang, Q.L, Liu, C.C. | | 登録日 | 2021-11-13 | | 公開日 | 2022-01-19 | | 最終更新日 | 2024-06-26 | | 実験手法 | ELECTRON MICROSCOPY (2.7 Å) | | 主引用文献 | Molecular basis of differential receptor usage for naturally occurring CD55-binding and -nonbinding coxsackievirus B3 strains.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7VY6
 
 | | Coxsackievirus B3(VP3-234N) incubate with CD55 at pH7.4 | | 分子名称: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3, ... | | 著者 | Wang, Q.L, Liu, C.C. | | 登録日 | 2021-11-13 | | 公開日 | 2022-01-19 | | 最終更新日 | 2022-08-10 | | 実験手法 | ELECTRON MICROSCOPY (3.02 Å) | | 主引用文献 | Molecular basis of differential receptor usage for naturally occurring CD55-binding and -nonbinding coxsackievirus B3 strains.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7W14
 
 | |
7W17
 
 | |
7YTJ
 
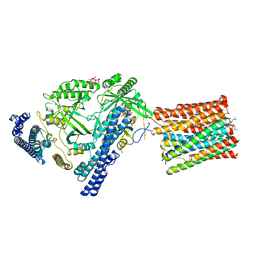 | | Cryo-EM structure of VTC complex | | 分子名称: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, INOSITOL HEXAKISPHOSPHATE, PHOSPHATE ION, ... | | 著者 | Guan, Z.Y, Chen, J, Liu, R.W, Chen, Y.K, Xing, Q, Du, Z.M, Liu, Z. | | 登録日 | 2022-08-15 | | 公開日 | 2023-02-22 | | 最終更新日 | 2024-07-03 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | The cytoplasmic synthesis and coupled membrane translocation of eukaryotic polyphosphate by signal-activated VTC complex.
Nat Commun, 14, 2023
|
|
6K0B
 
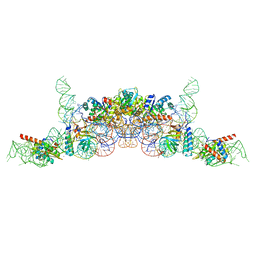 | | cryo-EM structure of archaeal Ribonuclease P with mature tRNA | | 分子名称: | 50S ribosomal protein L7Ae, RPR, Ribonuclease P protein component 1, ... | | 著者 | Wan, F, Lan, P, Wu, J, Lei, M. | | 登録日 | 2019-05-05 | | 公開日 | 2019-06-19 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (4.3 Å) | | 主引用文献 | Cryo-electron microscopy structure of an archaeal ribonuclease P holoenzyme.
Nat Commun, 10, 2019
|
|
6K0A
 
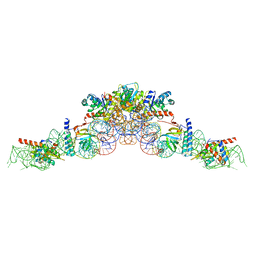 | | cryo-EM structure of an archaeal Ribonuclease P | | 分子名称: | 50S ribosomal protein L7Ae, RPR, Ribonuclease P protein component 1, ... | | 著者 | Wan, F, Lan, P, Wu, J, Lei, M. | | 登録日 | 2019-05-05 | | 公開日 | 2019-06-19 | | 最終更新日 | 2022-12-21 | | 実験手法 | ELECTRON MICROSCOPY (4.6 Å) | | 主引用文献 | Cryo-electron microscopy structure of an archaeal ribonuclease P holoenzyme.
Nat Commun, 10, 2019
|
|
3TJC
 
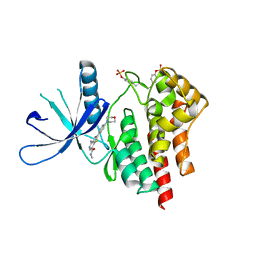 | | Co-crystal structure of jak2 with thienopyridine 8 | | 分子名称: | 4-amino-N-methyl-2-[4-(morpholin-4-yl)phenyl]thieno[3,2-c]pyridine-7-carboxamide, Tyrosine-protein kinase JAK2 | | 著者 | Huang, X. | | 登録日 | 2011-08-24 | | 公開日 | 2011-11-30 | | 最終更新日 | 2011-12-28 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Discovery of potent and highly selective thienopyridine janus kinase 2 inhibitors.
J.Med.Chem., 54, 2011
|
|
3TJD
 
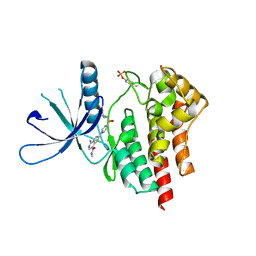 | | co-crystal structure of Jak2 with thienopyridine 19 | | 分子名称: | 4-amino-2-[4-(tert-butylsulfamoyl)phenyl]-N-methylthieno[3,2-c]pyridine-7-carboxamide, Tyrosine-protein kinase JAK2 | | 著者 | Huang, X. | | 登録日 | 2011-08-24 | | 公開日 | 2011-11-30 | | 最終更新日 | 2011-12-28 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Discovery of potent and highly selective thienopyridine janus kinase 2 inhibitors.
J.Med.Chem., 54, 2011
|
|
4WSI
 
 | | Crystal Structure of PALS1/Crb complex | | 分子名称: | MAGUK p55 subfamily member 5, Protein crumbs | | 著者 | Wei, Z, Li, Y, Zhang, M. | | 登録日 | 2014-10-28 | | 公開日 | 2014-11-26 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.95 Å) | | 主引用文献 | Structure of Crumbs tail in complex with the PALS1 PDZ-SH3-GK tandem reveals a highly specific assembly mechanism for the apical Crumbs complex.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
8HOG
 
 | | Crystal structure of Bcl-2 in complex with sonrotoclax | | 分子名称: | Apoptosis regulator Bcl-2, ~{N}-[4-[(4-methyl-4-oxidanyl-cyclohexyl)methylamino]-3-nitro-phenyl]sulfonyl-4-[2-[(2~{S})-2-(2-propan-2-ylphenyl)pyrrolidin-1-yl]-7-azaspiro[3.5]nonan-7-yl]-2-(1~{H}-pyrrolo[2,3-b]pyridin-5-yloxy)benzamide | | 著者 | Liu, J, Xu, M, Feng, Y, Hong, Y, Liu, Y. | | 登録日 | 2022-12-10 | | 公開日 | 2024-01-17 | | 最終更新日 | 2024-05-15 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Sonrotoclax overcomes BCL2 G101V mutation-induced venetoclax resistance in preclinical models of hematologic malignancy.
Blood, 143, 2024
|
|
8HOH
 
 | | Crystal structure of Bcl-2 G101V in complex with sonrotoclax | | 分子名称: | Apoptosis regulator Bcl-2, ~{N}-[4-[(4-methyl-4-oxidanyl-cyclohexyl)methylamino]-3-nitro-phenyl]sulfonyl-4-[2-[(2~{S})-2-(2-propan-2-ylphenyl)pyrrolidin-1-yl]-7-azaspiro[3.5]nonan-7-yl]-2-(1~{H}-pyrrolo[2,3-b]pyridin-5-yloxy)benzamide | | 著者 | Liu, J, Xu, M, Feng, Y, Hong, Y, Liu, Y. | | 登録日 | 2022-12-10 | | 公開日 | 2024-01-17 | | 最終更新日 | 2024-05-15 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Sonrotoclax overcomes BCL2 G101V mutation-induced venetoclax resistance in preclinical models of hematologic malignancy.
Blood, 143, 2024
|
|
8HOI
 
 | | Crystal structure of Bcl-2 D103Y in complex with sonrotoclax | | 分子名称: | Apoptosis regulator Bcl-2, FORMIC ACID, ~{N}-[4-[(4-methyl-4-oxidanyl-cyclohexyl)methylamino]-3-nitro-phenyl]sulfonyl-4-[2-[(2~{S})-2-(2-propan-2-ylphenyl)pyrrolidin-1-yl]-7-azaspiro[3.5]nonan-7-yl]-2-(1~{H}-pyrrolo[2,3-b]pyridin-5-yloxy)benzamide | | 著者 | Liu, J, Xu, M, Feng, Y, Hong, Y, Liu, Y. | | 登録日 | 2022-12-10 | | 公開日 | 2024-01-17 | | 最終更新日 | 2024-05-15 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Sonrotoclax overcomes BCL2 G101V mutation-induced venetoclax resistance in preclinical models of hematologic malignancy.
Blood, 143, 2024
|
|
5J1A
 
 | | Antigen presenting molecule | | 分子名称: | 3-[(8Z,11Z)-pentadeca-8,11-dien-1-yl]benzene-1,2-diol, Beta-2-microglobulin, T-cell surface glycoprotein CD1a | | 著者 | Yongqing, T, Rossjohn, J, Le Nours, J, Marquez, E, Winau, F. | | 登録日 | 2016-03-29 | | 公開日 | 2016-08-03 | | 最終更新日 | 2018-04-04 | | 実験手法 | X-RAY DIFFRACTION (1.86 Å) | | 主引用文献 | CD1a on Langerhans cells controls inflammatory skin disease.
Nat. Immunol., 17, 2016
|
|
3DAU
 
 | |
3DAT
 
 | |
7V6Y
 
 | | Cryo-EM structure of Patched in lipid nanodisc - the wildtype, 3.5 angstrom (re-processed with dataset of 7dzq) | | 分子名称: | (2S)-2-azanyl-3-[[(2S)-3-butanoyloxy-2-dec-9-enoyloxy-propoxy]-oxidanyl-phosphoryl]oxy-propanoic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Luo, Y, Zhao, Y, Qu, Q, Li, D. | | 登録日 | 2021-08-20 | | 公開日 | 2021-09-22 | | 最終更新日 | 2022-02-16 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | Cryo-EM study of patched in lipid nanodisc suggests a structural basis for its clustering in caveolae.
Structure, 29, 2021
|
|
7V6Z
 
 | | Cryo-EM structure of Patched1 (V1084A mutant) in lipid nanodisc, 3.64 angstrom (reprocessed with the dataset of 7dzp) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL, ... | | 著者 | Luo, Y, Zhao, Y, Qu, Q, Li, D. | | 登録日 | 2021-08-20 | | 公開日 | 2021-09-22 | | 最終更新日 | 2022-02-16 | | 実験手法 | ELECTRON MICROSCOPY (3.64 Å) | | 主引用文献 | Cryo-EM study of patched in lipid nanodisc suggests a structural basis for its clustering in caveolae.
Structure, 29, 2021
|
|
