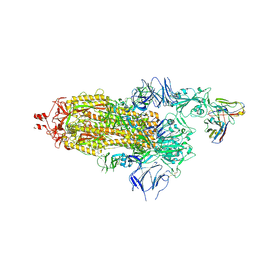
1OGA
 
 | | A structural basis for immunodominant human T-cell receptor recognition. | | 分子名称: | BETA-2-MICROGLOBULIN, GILGFVFTL, HLA CLASS I HISTOCOMPATIBILITY ANTIGEN, ... | | 著者 | Stewart-Jones, G.B.E, McMichael, A.J, Bell, J.I, Stuart, D.I, Jones, E.Y. | | 登録日 | 2003-04-28 | | 公開日 | 2003-06-19 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | A Structural Basis for Immunodominant Human T Cell Receptor Recognition
Nat.Immunol., 4, 2003
|
|
7PS6
 
 | | Crystal structure of the receptor binding domain of SARS-CoV-2 beta variant spike glycoprotein in complex with Beta-44 and Beta-54 Fabs | | 分子名称: | 2-(2-METHOXYETHOXY)ETHANOL, Beta-44 Fab heavy chain, Beta-44 Fab light chain, ... | | 著者 | Zhou, D, Ren, J, Stuart, D.I. | | 登録日 | 2021-09-22 | | 公開日 | 2021-12-15 | | 最終更新日 | 2024-11-13 | | 実験手法 | X-RAY DIFFRACTION (2.26 Å) | | 主引用文献 | The antibody response to SARS-CoV-2 Beta underscores the antigenic distance to other variants.
Cell Host Microbe, 30, 2022
|
|
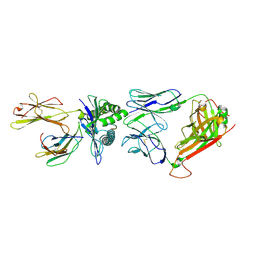
7Q0G
 
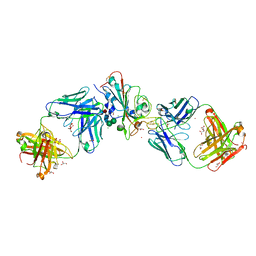 | | Crystal structure of the receptor binding domain of SARS-CoV-2 beta variant spike glycoprotein in complex with Beta-49 and FI-3A Fabs | | 分子名称: | Beta-49 Fab heavy chain, Beta-49 Fab light chain, CHLORIDE ION, ... | | 著者 | Zhou, D, Ren, J, Stuart, D.I. | | 登録日 | 2021-10-14 | | 公開日 | 2021-12-22 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (1.82 Å) | | 主引用文献 | The antibody response to SARS-CoV-2 Beta underscores the antigenic distance to other variants.
Cell Host Microbe, 30, 2022
|
|
7Q0H
 
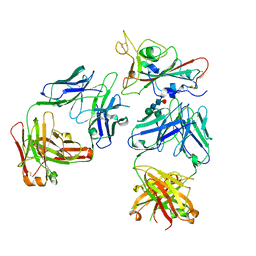 | | Crystal structure of the receptor binding domain of SARS-CoV-2 beta variant spike glycoprotein in complex with Beta-50 and Beta-54 | | 分子名称: | Beta-50 Fab heavy chain, Beta-50 Fab light chain, Beta-54 Fab heavy chain, ... | | 著者 | Zhou, D, Ren, J, Stuart, D.I. | | 登録日 | 2021-10-14 | | 公開日 | 2021-12-22 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (3.65 Å) | | 主引用文献 | The antibody response to SARS-CoV-2 Beta underscores the antigenic distance to other variants.
Cell Host Microbe, 30, 2022
|
|
7Q0I
 
 | | Crystal structure of the N-terminal domain of SARS-CoV-2 beta variant spike glycoprotein in complex with Beta-43 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Zhou, D, Ren, J, Stuart, D.I. | | 登録日 | 2021-10-14 | | 公開日 | 2021-12-22 | | 最終更新日 | 2024-11-13 | | 実験手法 | X-RAY DIFFRACTION (2.39 Å) | | 主引用文献 | The antibody response to SARS-CoV-2 Beta underscores the antigenic distance to other variants.
Cell Host Microbe, 30, 2022
|
|
7Q0A
 
 | | SARS-CoV-2 Spike ectodomain with Fab FI3A | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, FI3A fab Light chain, ... | | 著者 | Duyvesteyn, H.M.E, Ren, J, Stuart, D.I. | | 登録日 | 2021-10-14 | | 公開日 | 2022-02-23 | | 最終更新日 | 2024-11-06 | | 実験手法 | ELECTRON MICROSCOPY (4.8 Å) | | 主引用文献 | Structures and therapeutic potential of anti-RBD human monoclonal antibodies against SARS-CoV-2.
Theranostics, 12, 2022
|
|
8CIF
 
 | | Bovine naive ultralong antibody AbD08 collected at 293K | | 分子名称: | Heavy chain, Light chain | | 著者 | Clarke, J.D, Mikolajek, H, Stuart, D.I, Owens, R.J. | | 登録日 | 2023-02-09 | | 公開日 | 2023-05-24 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Protein-to-structure pipeline for ambient-temperature in situ crystallography at VMXi.
Iucrj, 10, 2023
|
|
8R1C
 
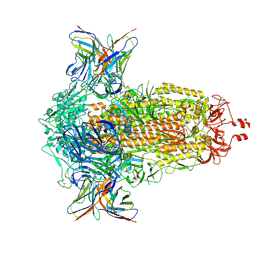 | | SD1-2 Fab in complex with SARS-CoV-2 BA.2.12.1 Spike Glycoprotein | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, SD1-2 fab heavy chain, SD1-2 fab light chain, ... | | 著者 | Duyvesteyn, H.M.E, Ren, J, Stuart, D.I. | | 登録日 | 2023-11-01 | | 公開日 | 2024-03-13 | | 最終更新日 | 2024-11-06 | | 実験手法 | ELECTRON MICROSCOPY (2.2 Å) | | 主引用文献 | The SARS-CoV-2 neutralizing antibody response to SD1 and its evasion by BA.2.86.
Nat Commun, 15, 2024
|
|
8R1D
 
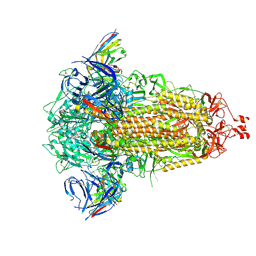 | | SD1-3 Fab in complex with SARS-CoV-2 BA.2.12.1 Spike Glycoprotein | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, SD1-3 Fab Heavy Chain, SD1-3 Fab Light Chain, ... | | 著者 | Duyvesteyn, H.M.E, Ren, J, Stuart, D.I. | | 登録日 | 2023-11-01 | | 公開日 | 2024-03-13 | | 最終更新日 | 2024-10-16 | | 実験手法 | ELECTRON MICROSCOPY (2.37 Å) | | 主引用文献 | The SARS-CoV-2 neutralizing antibody response to SD1 and its evasion by BA.2.86.
Nat Commun, 15, 2024
|
|
8R8K
 
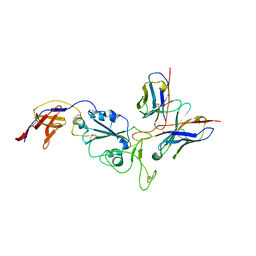 | | XBB-4 Fab in complex with SARS-CoV-2 BA.2.12.1 Spike Glycoprotein | | 分子名称: | Spike glycoprotein,Fibritin, XBB-4 Fab Heavy chain, XBB-4 Fab Light chain | | 著者 | Duyvesteyn, H.M.E, Ren, J, Stuart, D.I. | | 登録日 | 2023-11-29 | | 公開日 | 2024-05-08 | | 最終更新日 | 2024-11-13 | | 実験手法 | ELECTRON MICROSCOPY (3.41 Å) | | 主引用文献 | A structure-function analysis shows SARS-CoV-2 BA.2.86 balances antibody escape and ACE2 affinity.
Cell Rep Med, 5, 2024
|
|
4ZQX
 
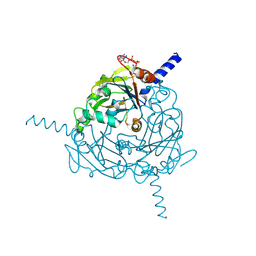 | | A revised partiality model and post-refinement algorithm for X-ray free-electron laser data | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Polyhedrin | | 著者 | Ginn, H.M, Brewster, A.S, Hattne, J, Evans, G, Wagner, A, Grimes, J, Sauter, N.K, Sutton, G, Stuart, D.I. | | 登録日 | 2015-05-11 | | 公開日 | 2015-06-03 | | 最終更新日 | 2024-11-20 | | 実験手法 | X-RAY DIFFRACTION (1.46 Å) | | 主引用文献 | A revised partiality model and post-refinement algorithm for X-ray free-electron laser data.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
5A9B
 
 | | Crystal structure of Bombyx mori CPV1 polyhedra base domain deleted mutant | | 分子名称: | POLYHEDRIN | | 著者 | Ji, X, Axford, D, Owen, R, Evans, G, Ginn, H.M, Sutton, G, Stuart, D.I. | | 登録日 | 2015-07-17 | | 公開日 | 2015-09-02 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.883 Å) | | 主引用文献 | Polyhedra Structures and the Evolution of the Insect Viruses.
J.Struct.Biol., 192, 2015
|
|
5A9C
 
 | | Crystal structure of Antheraea mylitta CPV4 polyhedra base domain deleted mutant | | 分子名称: | POLYHEDRIN | | 著者 | Ji, X, Axford, D, Owen, R, Evans, G, Ginn, H.M, Sutton, G, Stuart, D.I. | | 登録日 | 2015-07-17 | | 公開日 | 2015-09-02 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (1.71 Å) | | 主引用文献 | Polyhedra Structures and the Evolution of the Insect Viruses.
J.Struct.Biol., 192, 2015
|
|
5A98
 
 | | Crystal structure of Trichoplusia ni CPV15 polyhedra | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, POLYHEDRIN | | 著者 | Ji, X, Axford, D, Owen, R, Evans, G, Ginn, H.M, Sutton, G, Stuart, D.I. | | 登録日 | 2015-07-17 | | 公開日 | 2015-09-02 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.816 Å) | | 主引用文献 | Polyhedra Structures and the Evolution of the Insect Viruses.
J.Struct.Biol., 192, 2015
|
|
5A96
 
 | | Crystal structure of Lymantria dispar CPV14 polyhedra | | 分子名称: | GUANOSINE-5'-TRIPHOSPHATE, POLYHEDRIN | | 著者 | Ji, X, Axford, D, Owen, R, Evans, G, Ginn, H.M, Sutton, G, Stuart, D.I. | | 登録日 | 2015-07-17 | | 公開日 | 2015-09-02 | | 最終更新日 | 2024-10-09 | | 実験手法 | X-RAY DIFFRACTION (1.914 Å) | | 主引用文献 | Polyhedra Structures and the Evolution of the Insect Viruses.
J.Struct.Biol., 192, 2015
|
|
5A99
 
 | | Crystal structure of Operophtera brumata CPV19 polyhedra | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Polyhedrin | | 著者 | Ji, X, Axford, D, Owen, R, Evans, G, Ginn, H.M, Sutton, G, Stuart, D.I. | | 登録日 | 2015-07-17 | | 公開日 | 2015-09-02 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (1.511 Å) | | 主引用文献 | Polyhedra Structures and the Evolution of the Insect Viruses.
J.Struct.Biol., 192, 2015
|
|
5A8U
 
 | | Crystal structure of Orgyia pseudotsugata CPV5 polyhedra | | 分子名称: | POLYHEDRIN | | 著者 | Ji, X, Axford, D, Owen, R, Evans, G, Ginn, H.M, Sutton, G, Stuart, D.I. | | 登録日 | 2015-07-17 | | 公開日 | 2015-09-02 | | 最終更新日 | 2024-11-13 | | 実験手法 | X-RAY DIFFRACTION (1.609 Å) | | 主引用文献 | Polyhedra Structures and the Evolution of the Insect Viruses.
J.Struct.Biol., 192, 2015
|
|
5ACA
 
 | | Structure-based energetics of protein interfaces guide Foot-and-Mouth disease virus vaccine design | | 分子名称: | VP1, VP2, VP3, ... | | 著者 | Kotecha, A, Seago, J, Scott, K, Burman, A, Loureiro, S, Ren, J, Porta, C, Ginn, H.M, Jackson, T, Perez-Martin, E, Siebert, C.A, Paul, G, Huiskonen, J.T, Jones, I.M, Esnouf, R.M, Fry, E.E, Maree, F.F, Charleston, B, Stuart, D.I. | | 登録日 | 2015-08-14 | | 公開日 | 2015-09-23 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | Structure-Based Energetics of Protein Interfaces Guide Foot-and-Mouth Disease Vaccine Design
Nat.Struct.Mol.Biol., 22, 2015
|
|
5A9A
 
 | | Crystal structure of Simulium ubiquitum CPV20 polyhedra | | 分子名称: | POLYHEDRIN, URIDINE 5'-TRIPHOSPHATE | | 著者 | Ji, X, Axford, D, Owen, R, Evans, G, Ginn, H.M, Sutton, G, Stuart, D.I. | | 登録日 | 2015-07-17 | | 公開日 | 2015-09-02 | | 最終更新日 | 2024-11-20 | | 実験手法 | X-RAY DIFFRACTION (1.822 Å) | | 主引用文献 | Polyhedra Structures and the Evolution of the Insect Viruses.
J.Struct.Biol., 192, 2015
|
|
5A8T
 
 | | Crystal structure of Antheraea mylitta CPV4 polyhedra type 2 | | 分子名称: | POLYHEDRIN | | 著者 | Ji, X, Axford, D, Owen, R, Evans, G, Ginn, H.M, Sutton, G, Stuart, D.I. | | 登録日 | 2015-07-17 | | 公開日 | 2015-09-02 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.003 Å) | | 主引用文献 | Polyhedra Structures and the Evolution of the Insect Viruses.
J.Struct.Biol., 192, 2015
|
|
5C4W
 
 | | Crystal structure of coxsackievirus A16 | | 分子名称: | CHLORIDE ION, POTASSIUM ION, SODIUM ION, ... | | 著者 | Ren, J, Wang, X, Zhu, L, Hu, Z, Gao, Q, Yang, P, Li, X, Wang, J, Shen, X, Fry, E.E, Rao, Z, Stuart, D.I. | | 登録日 | 2015-06-18 | | 公開日 | 2015-08-26 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.65 Å) | | 主引用文献 | Structures of Coxsackievirus A16 Capsids with Native Antigenicity: Implications for Particle Expansion, Receptor Binding, and Immunogenicity.
J.Virol., 89, 2015
|
|
5C9A
 
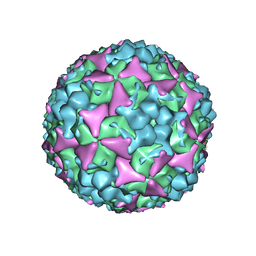 | | Crystal structure of empty coxsackievirus A16 particle | | 分子名称: | CHLORIDE ION, POTASSIUM ION, SPHINGOSINE, ... | | 著者 | Ren, J, Wang, X, Zhu, L, Hu, Z, Gao, Q, Yang, P, Li, X, Wang, J, Shen, X, Fry, E.E, Rao, Z, Stuart, D.I. | | 登録日 | 2015-06-26 | | 公開日 | 2015-08-26 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Structures of Coxsackievirus A16 Capsids with Native Antigenicity: Implications for Particle Expansion, Receptor Binding, and Immunogenicity.
J.Virol., 89, 2015
|
|
5A8V
 
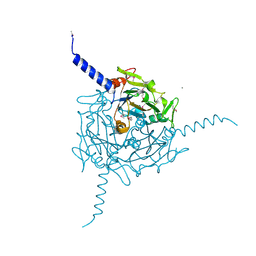 | | Crystal structure of Orgyia pseudotsugata CPV5 polyhedra with SeMet substitution | | 分子名称: | CALCIUM ION, POLYHEDRIN | | 著者 | Ji, X, Axford, D, Owen, R, Evans, G, Ginn, H.M, Sutton, G, Stuart, D.I. | | 登録日 | 2015-07-17 | | 公開日 | 2015-09-02 | | 最終更新日 | 2024-11-20 | | 実験手法 | X-RAY DIFFRACTION (2.074 Å) | | 主引用文献 | Polyhedra Structures and the Evolution of the Insect Viruses.
J.Struct.Biol., 192, 2015
|
|
5A9P
 
 | | Crystal structure of Operophtera brumata CPV18 polyhedra | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, POLYHEDRIN | | 著者 | Ji, X, Axford, D, Owen, R, Evans, G, Ginn, H.M, Sutton, G, Stuart, D.I. | | 登録日 | 2015-07-21 | | 公開日 | 2015-09-02 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.476 Å) | | 主引用文献 | Polyhedra Structures and the Evolution of the Insect Viruses.
J.Struct.Biol., 192, 2015
|
|
5ABJ
 
 | | Structure of Coxsackievirus A16 in complex with GPP3 | | 分子名称: | 1-[(3S)-5-[4-[(E)-ETHOXYIMINOMETHYL]PHENOXY]-3-METHYL-PENTYL]-3-PYRIDIN-4-YL-IMIDAZOLIDIN-2-ONE, CHLORIDE ION, SODIUM ION, ... | | 著者 | De Colibus, L, Wang, X, Tijsma, A, Neyts, J, Spyrou, J.A.B, Ren, J, Grimes, J.M, Puerstinger, G, Leyssen, P, Fry, E.E, Rao, Z, Stuart, D.I. | | 登録日 | 2015-08-06 | | 公開日 | 2015-09-09 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.75 Å) | | 主引用文献 | Structure Elucidation of Coxsackievirus A16 in Complex with Gpp3 Informs a Systematic Review of Highly Potent Capsid Binders to Enteroviruses.
Plos Pathog., 11, 2015
|
|
