7WA1
 
 | |
8Y9C
 
 | |
8Y9B
 
 | | TcdB1 in complex with mini-binder | | 分子名称: | De novo design mini-binder, Toxin B, ZINC ION | | 著者 | Lv, X.C, Lu, P.L. | | 登録日 | 2024-02-06 | | 公開日 | 2024-08-28 | | 最終更新日 | 2024-10-09 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | De novo design of mini-protein binders broadly neutralizing Clostridioides difficile toxin B variants
Nat Commun, 2024
|
|
6H8P
 
 | | JMJD2A/ KDM4A COMPLEXED WITH NI(II), NOG AND Histone H1.4(18-32)K26me3 peptide (15-mer) | | 分子名称: | CHLORIDE ION, GLYCEROL, Histone H1.4, ... | | 著者 | Chowdhury, R, Walport, L.J, Schofield, C.J. | | 登録日 | 2018-08-03 | | 公開日 | 2018-08-15 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.983 Å) | | 主引用文献 | Mechanistic and structural studies of KDM-catalysed demethylation of histone 1 isotype 4 at lysine 26.
FEBS Lett., 592, 2018
|
|
7V2Y
 
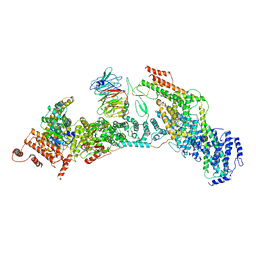 | | cryo-EM structure of yeast THO complex with Sub2 | | 分子名称: | ATP-dependent RNA helicase SUB2, Protein TEX1, THO complex subunit 2, ... | | 著者 | Chen, C, Tan, M, Wu, Z, Wu, J, Lei, M. | | 登録日 | 2021-08-10 | | 公開日 | 2022-07-27 | | 最終更新日 | 2024-06-12 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Structural and functional insights into R-loop prevention and mRNA export by budding yeast THO-Sub2 complex.
Sci Bull (Beijing), 66, 2021
|
|
7V2W
 
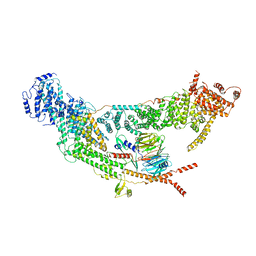 | | protomer structure from the dimer of yeast THO complex | | 分子名称: | Protein TEX1, THO complex subunit 2, THO complex subunit HPR1, ... | | 著者 | Chen, C, Tan, M, Wu, Z, Wu, J, Lei, M. | | 登録日 | 2021-08-10 | | 公開日 | 2022-07-27 | | 最終更新日 | 2024-06-12 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Structural and functional insights into R-loop prevention and mRNA export by budding yeast THO-Sub2 complex.
Sci Bull (Beijing), 66, 2021
|
|
6JW1
 
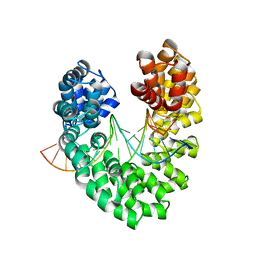 | | Universal RVD R* accommodates 5mC via water-mediated interactions | | 分子名称: | DNA (5'-D(*AP*GP*AP*GP*AP*CP*GP*CP*GP*AP*AP*GP*GP*GP*AP*CP*A)-3'), DNA (5'-D(*TP*GP*TP*CP*CP*CP*TP*TP*(5CM)P*GP*CP*GP*TP*CP*TP*CP*T)-3'), TAL effector | | 著者 | Liu, L, Yi, C. | | 登録日 | 2019-04-18 | | 公開日 | 2020-04-22 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.49 Å) | | 主引用文献 | Structural Insights into the Specific Recognition of 5-methylcytosine and 5-hydroxymethylcytosine by TAL Effectors.
J.Mol.Biol., 432, 2020
|
|
7VNI
 
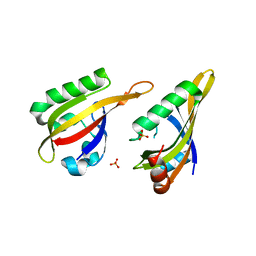 | | AHR-ARNT PAS-B heterodimer | | 分子名称: | Ahr homolog spineless, Aryl hydrocarbon receptor nuclear translocator, SULFATE ION | | 著者 | Dai, S.Y. | | 登録日 | 2021-10-11 | | 公開日 | 2022-04-13 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.997 Å) | | 主引用文献 | Structural insight into the ligand binding mechanism of aryl hydrocarbon receptor.
Nat Commun, 13, 2022
|
|
7VNA
 
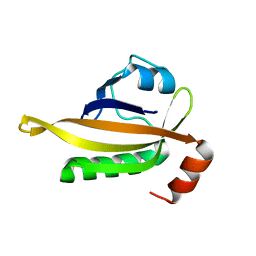 | | drosophlia AHR PAS-B domain | | 分子名称: | Ahr homolog spineless | | 著者 | Dai, S.Y. | | 登録日 | 2021-10-10 | | 公開日 | 2022-04-13 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.597 Å) | | 主引用文献 | Structural insight into the ligand binding mechanism of aryl hydrocarbon receptor.
Nat Commun, 13, 2022
|
|
7VNH
 
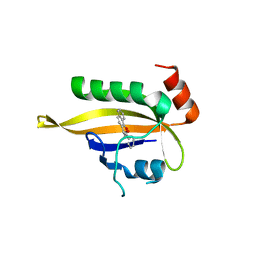 | |
7VM0
 
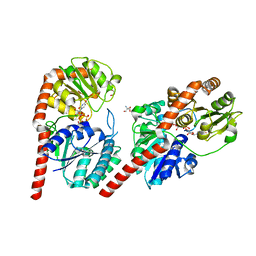 | | Crystal structure of YojK from B.subtilis in complex with UDP | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, Glycosyl transferase family 1, ... | | 著者 | Hou, X.D, Guo, B.D, Rao, Y.J. | | 登録日 | 2021-10-06 | | 公開日 | 2022-10-12 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Highly efficient production of rebaudioside D enabled by structure-guided engineering of bacterial glycosyltransferase YojK.
Front Bioeng Biotechnol, 10, 2022
|
|
6JW2
 
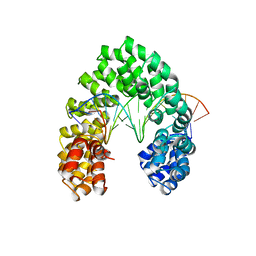 | | Universal RVD R* accommodates 5hmC via water-mediated interactions | | 分子名称: | DNA (5'-D(*AP*GP*AP*GP*AP*CP*GP*CP*GP*AP*AP*GP*GP*GP*AP*CP*A)-3'), DNA (5'-D(*TP*GP*TP*CP*CP*CP*TP*TP*(5HC)P*GP*CP*GP*TP*CP*TP*CP*T)-3'), TAL effector | | 著者 | Liu, L, Yi, C. | | 登録日 | 2019-04-18 | | 公開日 | 2020-04-22 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (3.03 Å) | | 主引用文献 | Structural Insights into the Specific Recognition of 5-methylcytosine and 5-hydroxymethylcytosine by TAL Effectors.
J.Mol.Biol., 432, 2020
|
|
7CH0
 
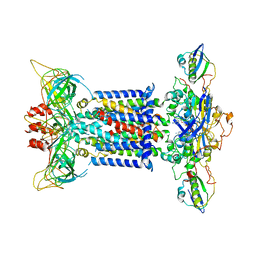 | | The overall structure of the MlaFEDB complex in ATP-bound EQclose conformation (Mutation of E170Q on MlaF) | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, Lipid asymmetry maintenance ABC transporter permease subunit MlaE, Lipid asymmetry maintenance protein MlaB, ... | | 著者 | Chi, X.M, Fan, Q.X, Zhang, Y.Y, Liang, K, Zhou, Q, Li, Y.Y. | | 登録日 | 2020-07-03 | | 公開日 | 2020-09-09 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Structural mechanism of phospholipids translocation by MlaFEDB complex.
Cell Res., 30, 2020
|
|
6JVZ
 
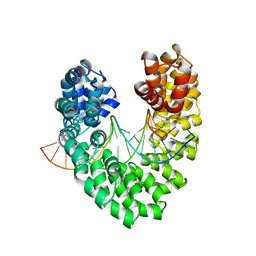 | | RVD HA specifically contacts 5mC through van der Waals interactions | | 分子名称: | DNA (5'-D(*AP*GP*AP*GP*AP*CP*GP*CP*GP*AP*AP*GP*GP*GP*AP*CP*A)-3'), DNA (5'-D(*TP*GP*TP*CP*CP*CP*TP*TP*(5CM)P*GP*CP*GP*TP*CP*TP*CP*T)-3'), TAL effector | | 著者 | Liu, L, Yi, C. | | 登録日 | 2019-04-18 | | 公開日 | 2020-04-22 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.48 Å) | | 主引用文献 | Structural Insights into the Specific Recognition of 5-methylcytosine and 5-hydroxymethylcytosine by TAL Effectors.
J.Mol.Biol., 432, 2020
|
|
7CGN
 
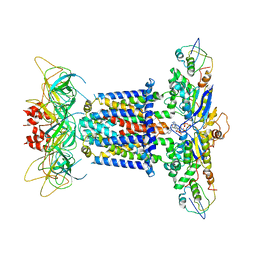 | | The overall structure of the MlaFEDB complex in ATP-bound EQtall conformation (Mutation of E170Q on MlaF) | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, Lipid asymmetry maintenance ABC transporter permease subunit MlaE, Lipid asymmetry maintenance protein MlaB, ... | | 著者 | Chi, X.M, Fan, Q.X, Zhang, Y.Y, Liang, K, Zhou, Q, Li, Y.Y. | | 登録日 | 2020-07-01 | | 公開日 | 2020-09-09 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (4.3 Å) | | 主引用文献 | Structural mechanism of phospholipids translocation by MlaFEDB complex.
Cell Res., 30, 2020
|
|
7CGE
 
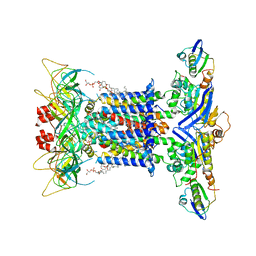 | | The overall structure of nucleotide free MlaFEDB complex | | 分子名称: | (1R)-2-{[(S)-{[(2S)-2,3-dihydroxypropyl]oxy}(hydroxy)phosphoryl]oxy}-1-[(hexadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate, Lipid asymmetry maintenance ABC transporter permease subunit MlaE, Lipid asymmetry maintenance protein MlaB, ... | | 著者 | Chi, X.M, Fan, Q.X, Zhang, Y.Y, Liang, K, Zhou, Q, Li, Y.Y. | | 登録日 | 2020-07-01 | | 公開日 | 2020-09-09 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | Structural mechanism of phospholipids translocation by MlaFEDB complex.
Cell Res., 30, 2020
|
|
6K62
 
 | |
3C2T
 
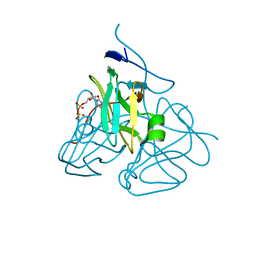 | | Evolution of chlorella virus dUTPase | | 分子名称: | DEOXYURIDINE-5'-DIPHOSPHATE, Deoxyuridine triphosphatase, MAGNESIUM ION | | 著者 | Yamanishi, M, Homma, K, Zhang, Y, Etten, L.V.J, Moriyama, H. | | 登録日 | 2008-01-25 | | 公開日 | 2009-02-24 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Crystallization and crystal-packing studies of Chlorella virus deoxyuridine triphosphatase.
Acta Crystallogr.,Sect.F, 65, 2009
|
|
7ROM
 
 | |
3FE5
 
 | | Crystal structure of 3-hydroxyanthranilate 3,4-dioxygenase from bovine kidney | | 分子名称: | 3-hydroxyanthranilate 3,4-dioxygenase, FE (III) ION | | 著者 | Dilovic, I, Gliubich, F, Malpeli, G, Zanotti, G, Matkovic-Calogovic, D. | | 登録日 | 2008-11-27 | | 公開日 | 2009-06-09 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.51 Å) | | 主引用文献 | Crystal structure of bovine 3-hydroxyanthranilate 3,4-dioxygenase.
Biopolymers, 2009
|
|
5Y1H
 
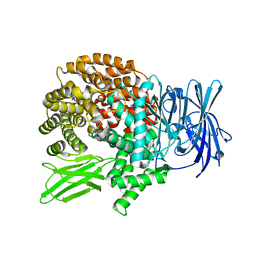 | | Crystal structure of Plasmodium falciparum aminopeptidase N in complex with (S)-2-(3-(2,4-difluorobenzyl)ureido)-N-hydroxy-4-methylpentanamide | | 分子名称: | (2R)-2-[[2,4-bis(fluoranyl)phenyl]methylcarbamoylamino]-4-methyl-N-oxidanyl-pentanamide, GLYCEROL, M1 family aminopeptidase, ... | | 著者 | Marapaka, A.K, Zhang, Y, Addlagatta, A. | | 登録日 | 2017-07-20 | | 公開日 | 2018-08-01 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.73 Å) | | 主引用文献 | Crystal structure of Plasmodium falciparum aminopeptidase N in complex with (S)-2-(3-(2,4-difluorobenzyl)ureido)-N-hydroxy-4-methylpentanamide
To Be Published
|
|
5Y1R
 
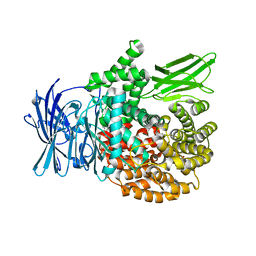 | |
5Y3I
 
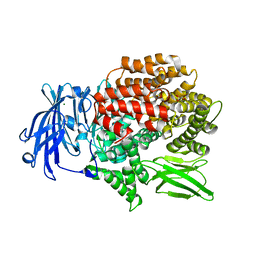 | |
5Y1S
 
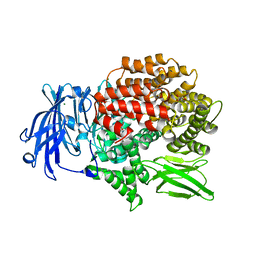 | | Crystal structure of Plasmodium falciparum aminopeptidase N in complex with (S)-2-(3-(3,4-dimethylbenzyl)ureido)-N-hydroxy-4-methylpentanamide | | 分子名称: | (2R)-2-[(3,4-dimethylphenyl)methylcarbamoylamino]-4-methyl-N-oxidanyl-pentanamide, (2S)-2-[(3,4-dimethylphenyl)methylcarbamoylamino]-4-methyl-N-oxidanyl-pentanamide, GLYCEROL, ... | | 著者 | Marapaka, A.K, Zhang, Y, Addlagatta, A. | | 登録日 | 2017-07-21 | | 公開日 | 2018-08-01 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.66 Å) | | 主引用文献 | Crystal structure of Plasmodium falciparum aminopeptidase N in complex with (S)-2-(3-(3,4-dimethylbenzyl)ureido)-N-hydroxy-4-methylpentanamide
To Be Published
|
|
5Y1V
 
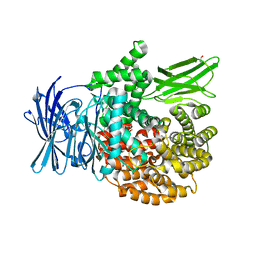 | | Crystal structure of Plasmodium falciparum aminopeptidase N in complex with (S)-2-(3-(2,6-diethylphenyl)ureido)- N-hydroxy-4-methylpentanamide | | 分子名称: | (2R)-2-[(2,6-diethylphenyl)carbamoylamino]-4-methyl-N-oxidanyl-pentanamide, GLYCEROL, M1 family aminopeptidase, ... | | 著者 | Marapaka, A.K, Zhang, Y, Addlagatta, A. | | 登録日 | 2017-07-21 | | 公開日 | 2018-08-01 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.62 Å) | | 主引用文献 | Crystal structure of Plasmodium falciparum aminopeptidase N in complex with (S)-2-(3-(2,6-diethylphenyl)ureido)- N-hydroxy-4-methylpentanamide
To Be Published
|
|
