6BR9
 
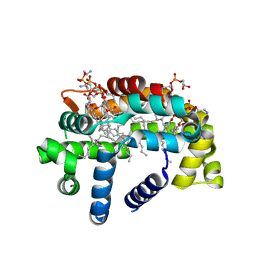 | | Structure of A6 reveals a novel lipid transporter | | 分子名称: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, Protein A6 homolog, [(2~{R})-1-[2-azanylethoxy(oxidanyl)phosphoryl]oxy-3-hexadecanoyloxy-propan-2-yl] (~{Z})-octadec-9-enoate | | 著者 | Deng, J, Peng, S, Pathak, P. | | 登録日 | 2017-11-30 | | 公開日 | 2018-06-20 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Structure of a lipid-bound viral membrane assembly protein reveals a modality for enclosing the lipid bilayer.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
3H0Z
 
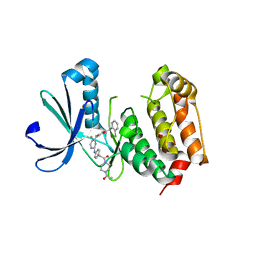 | | Aurora A in complex with a bisanilinopyrimidine | | 分子名称: | 4-{[2-({4-[2-(4-acetylpiperazin-1-yl)-2-oxoethyl]phenyl}amino)-5-fluoropyrimidin-4-yl]amino}-N-(2-chlorophenyl)benzamide, Serine/threonine-protein kinase 6 | | 著者 | Wiesmann, C, Ultsch, M.H, Cochran, A.G. | | 登録日 | 2009-04-10 | | 公開日 | 2009-07-07 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.92 Å) | | 主引用文献 | A class of 2,4-bisanilinopyrimidine Aurora A inhibitors with unusually high selectivity against Aurora B.
J.Med.Chem., 52, 2009
|
|
5WXH
 
 | | Crystal structure of TAF3 PHD finger bound to H3K4me3 | | 分子名称: | Histone H3K4me3, Transcription initiation factor TFIID subunit 3, ZINC ION | | 著者 | Zhao, S, Huang, J, Li, H. | | 登録日 | 2017-01-07 | | 公開日 | 2017-08-16 | | 最終更新日 | 2017-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.297 Å) | | 主引用文献 | Kinetic and high-throughput profiling of epigenetic interactions by 3D-carbene chip-based surface plasmon resonance imaging technology
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5WXG
 
 | | Structure of TAF PHD finger domain binds to H3(1-15)K4ac | | 分子名称: | Histone H3K4ac, MAGNESIUM ION, Transcription initiation factor TFIID subunit 3, ... | | 著者 | Zhao, S, Li, H. | | 登録日 | 2017-01-07 | | 公開日 | 2017-08-16 | | 最終更新日 | 2017-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.703 Å) | | 主引用文献 | Kinetic and high-throughput profiling of epigenetic interactions by 3D-carbene chip-based surface plasmon resonance imaging technology
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
3H10
 
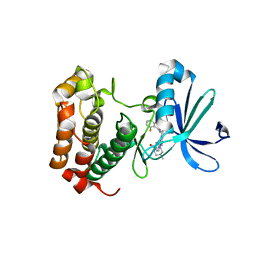 | | Aurora A inhibitor complex | | 分子名称: | 9-chloro-7-(2,6-difluorophenyl)-N-{4-[(4-methylpiperazin-1-yl)carbonyl]phenyl}-5H-pyrimido[5,4-d][2]benzazepin-2-amine, Serine/threonine-protein kinase 6 | | 著者 | Wiesmann, C, Ultsch, M.H, Cochran, A.G. | | 登録日 | 2009-04-10 | | 公開日 | 2009-07-07 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | A class of 2,4-bisanilinopyrimidine Aurora A inhibitors with unusually high selectivity against Aurora B.
J.Med.Chem., 52, 2009
|
|
3H0Y
 
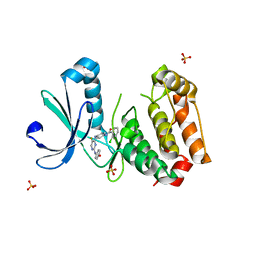 | | Aurora A in complex with a bisanilinopyrimidine | | 分子名称: | 2-chloro-N-[4-({5-fluoro-2-[(4-hydroxyphenyl)amino]pyrimidin-4-yl}amino)phenyl]benzamide, SULFATE ION, Serine/threonine-protein kinase 6 | | 著者 | Wiesmann, C, Ultsch, M.H, Cochran, A.G. | | 登録日 | 2009-04-10 | | 公開日 | 2009-07-07 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | A class of 2,4-bisanilinopyrimidine Aurora A inhibitors with unusually high selectivity against Aurora B.
J.Med.Chem., 52, 2009
|
|
6EDD
 
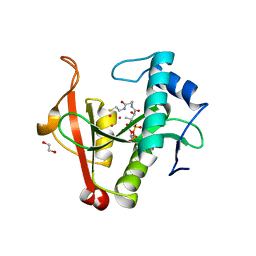 | | Crystal structure of a GNAT Superfamily PA3944 acetyltransferase in complex with CoA (P1 space group) | | 分子名称: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Acetyltransferase PA3944, ... | | 著者 | Czub, M.P, Porebski, P.J, Majorek, K.A, Satchell, K.J, Joachimiak, A, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2018-08-09 | | 公開日 | 2018-08-22 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | A Gcn5-Related N-Acetyltransferase (GNAT) Capable of Acetylating Polymyxin B and Colistin Antibiotics in Vitro.
Biochemistry, 57, 2018
|
|
6EDV
 
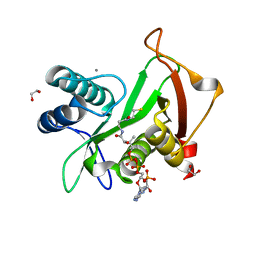 | | Structure of a GNAT superfamily acetyltransferase PA3944 in complex with CoA | | 分子名称: | 1,2-ETHANEDIOL, Acetyltransferase PA3944, CALCIUM ION, ... | | 著者 | Majorek, K.A, Satchell, K.J.F, Joachimiak, A, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2018-08-12 | | 公開日 | 2018-12-26 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.35 Å) | | 主引用文献 | A Gcn5-Related N-Acetyltransferase (GNAT) Capable of Acetylating Polymyxin B and Colistin Antibiotics in Vitro.
Biochemistry, 57, 2018
|
|
6E5P
 
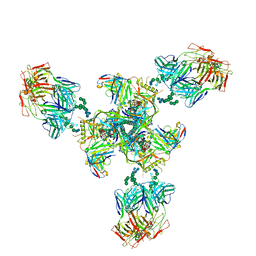 | | Backbone model based on cryo-EM map at 8.5 A of domain-swapped, glycan-reactive, neutralizing antibody 2G12 bound to HIV-1 Env BG505 DS-SOSIP, which was also bound to CD4-binding site antibody VRC03 | | 分子名称: | 2G12 Light chain, 2G12 heavy chain, Envelope glycoprotein gp120, ... | | 著者 | Acharya, P, Kwong, P.D. | | 登録日 | 2018-07-21 | | 公開日 | 2019-02-13 | | 最終更新日 | 2024-03-13 | | 実験手法 | ELECTRON MICROSCOPY (8.8 Å) | | 主引用文献 | Structural Survey of Broadly Neutralizing Antibodies Targeting the HIV-1 Env Trimer Delineates Epitope Categories and Characteristics of Recognition.
Structure, 27, 2019
|
|
4LST
 
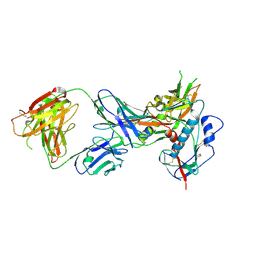 | | Crystal structure of broadly and potently neutralizing antibody VRC01 in complex with HIV-1 clade C strain ZM176.66 gp120 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ENVELOPE GLYCOPROTEIN GP120 of HIV-1 clade C, HEAVY CHAIN OF ANTIBODY VRC01, ... | | 著者 | Zhou, T, Moquin, S, Kwong, P.D. | | 登録日 | 2013-07-23 | | 公開日 | 2013-08-21 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (2.55 Å) | | 主引用文献 | Multidonor Analysis Reveals Structural Elements, Genetic Determinants, and Maturation Pathway for HIV-1 Neutralization by VRC01-Class Antibodies.
Immunity, 39, 2013
|
|
8RE4
 
 | |
8RED
 
 | |
8REE
 
 | |
8REC
 
 | |
8REA
 
 | |
8REB
 
 | |
6KYF
 
 | | Crystal structure of an anti-CRISPR protein | | 分子名称: | AcrF11, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | 著者 | Niu, Y, Wang, H, Zhang, Y, Feng, Y. | | 登録日 | 2019-09-18 | | 公開日 | 2020-09-23 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (3.07 Å) | | 主引用文献 | A Type I-F Anti-CRISPR Protein Inhibits the CRISPR-Cas Surveillance Complex by ADP-Ribosylation.
Mol.Cell, 80, 2020
|
|
6LAD
 
 | |
6LAF
 
 | | Crystal structure of the core domain of Amuc_1100 from Akkermansia muciniphila | | 分子名称: | Amuc_1100, SULFATE ION | | 著者 | Wang, J, Xiang, R, Zhang, M, Wang, M. | | 登録日 | 2019-11-12 | | 公開日 | 2020-08-05 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (3.001 Å) | | 主引用文献 | The variable oligomeric state of Amuc_1100 from Akkermansia muciniphila.
J.Struct.Biol., 212, 2020
|
|
4PNZ
 
 | | Human dipeptidyl peptidase IV/CD26 in complex with the long-acting inhibitor Omarigliptin (MK-3102) | | 分子名称: | (2R,3S,5R)-5-[2-(methylsulfonyl)-2,6-dihydropyrrolo[3,4-c]pyrazol-5(4H)-yl]-2-(2,4,5-trifluorophenyl)tetrahydro-2H-pyran-3-amine, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Scapin, G, Yan, Y. | | 登録日 | 2014-02-22 | | 公開日 | 2014-04-09 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Omarigliptin (MK-3102): A Novel Long-Acting DPP-4 Inhibitor for Once-Weekly Treatment of Type 2 Diabetes.
J.Med.Chem., 57, 2014
|
|
4IAN
 
 | | Crystal Structure of apo Human PRPF4B kinase domain | | 分子名称: | SULFATE ION, Serine/threonine-protein kinase PRP4 homolog | | 著者 | Mechin, I, Haas, K, Chen, X, Zhang, Y, McLean, L. | | 登録日 | 2012-12-06 | | 公開日 | 2013-08-28 | | 最終更新日 | 2017-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.44 Å) | | 主引用文献 | Evaluation of Cancer Dependence and Druggability of PRP4 Kinase Using Cellular, Biochemical, and Structural Approaches.
J.Biol.Chem., 288, 2013
|
|
4IFC
 
 | | Crystal Structure of ADP-bound Human PRPF4B kinase domain | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, SULFATE ION, Serine/threonine-protein kinase PRP4 homolog | | 著者 | Mechin, I, Haas, K, Chen, X, Zhang, Y, McLean, L. | | 登録日 | 2012-12-14 | | 公開日 | 2013-08-28 | | 最終更新日 | 2013-11-06 | | 実験手法 | X-RAY DIFFRACTION (2.13 Å) | | 主引用文献 | Evaluation of Cancer Dependence and Druggability of PRP4 Kinase Using Cellular, Biochemical, and Structural Approaches.
J.Biol.Chem., 288, 2013
|
|
4IJP
 
 | | Crystal Structure of Human PRPF4B kinase domain in complex with 4-{5-[(2-Chloro-pyridin-4-ylmethyl)-carbamoyl]-thiophen-2-yl}-benzo[b]thiophene-2-carboxylic acid amine | | 分子名称: | 4-(5-{[(2-chloropyridin-4-yl)methyl]carbamoyl}thiophen-2-yl)-1-benzothiophene-2-carboxamide, SULFATE ION, Serine/threonine-protein kinase PRP4 homolog | | 著者 | Mechin, I, Haas, K, Chen, X, Zhang, Y, McLean, L. | | 登録日 | 2012-12-22 | | 公開日 | 2013-08-28 | | 最終更新日 | 2017-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Evaluation of Cancer Dependence and Druggability of PRP4 Kinase Using Cellular, Biochemical, and Structural Approaches.
J.Biol.Chem., 288, 2013
|
|
6LZZ
 
 | | Crystal structure of the PDE9 catalytic domain in complex with inhibitor 4a | | 分子名称: | 1-cyclopentyl-6-[[(2R)-1-(2-oxa-6-azaspiro[3.3]heptan-6-yl)-1-oxidanylidene-propan-2-yl]amino]-5H-pyrazolo[3,4-d]pyrimidin-4-one, High affinity cGMP-specific 3',5'-cyclic phosphodiesterase 9A, MAGNESIUM ION, ... | | 著者 | Huang, Y.Y, Wu, Y, Luo, H.B. | | 登録日 | 2020-02-19 | | 公開日 | 2021-02-24 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.40003753 Å) | | 主引用文献 | Identification of phosphodiesterase-9 as a novel target for pulmonary arterial hypertension by using highly selective and orally bioavailable inhibitors
To Be Published
|
|
6M71
 
 | | SARS-Cov-2 RNA-dependent RNA polymerase in complex with cofactors | | 分子名称: | Non-structural protein 7, Non-structural protein 8, RNA-directed RNA polymerase | | 著者 | Gao, Y, Yan, L, Huang, Y, Liu, F, Cao, L, Wang, T, Wang, Q, Lou, Z, Rao, Z. | | 登録日 | 2020-03-16 | | 公開日 | 2020-04-01 | | 最終更新日 | 2021-03-10 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | Structure of the RNA-dependent RNA polymerase from COVID-19 virus.
Science, 368, 2020
|
|
