7LWK
 
 | |
7LWP
 
 | |
7LWN
 
 | |
4J6D
 
 | |
4J6B
 
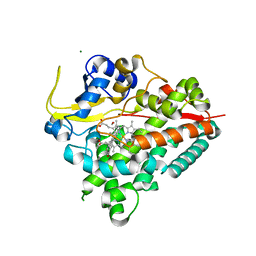 | |
4J6C
 
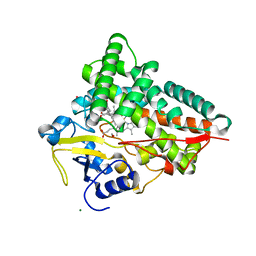 | |
4JBT
 
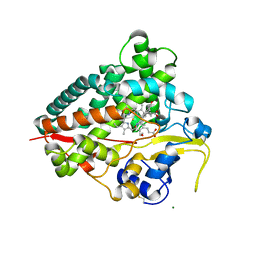 | |
3L9B
 
 | |
8U1D
 
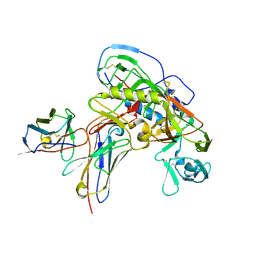 | | Cryo-EM structure of vaccine-elicited CD4 binding site antibody DH1285 bound to HIV-1 CH505TFchim.6R.SOSIP.664v4.1 Env Local Refinement | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, DH1285 Heavy Chain, DH1285 Light Chain, ... | | 著者 | Thakur, B, Stalls, V.D, Acharya, P. | | 登録日 | 2023-08-31 | | 公開日 | 2024-01-03 | | 最終更新日 | 2024-07-17 | | 実験手法 | ELECTRON MICROSCOPY (4.25 Å) | | 主引用文献 | Vaccine induction of CD4-mimicking HIV-1 broadly neutralizing antibody precursors in macaques.
Cell, 187, 2024
|
|
5CSZ
 
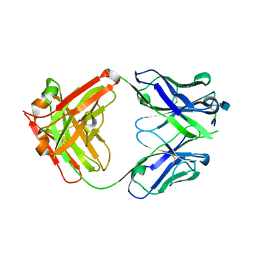 | | CRYSTAL STRUCTURE OF GANTENERUMAB FAB FRAGMENT IN COMPLEX WITH ABETA 1-11 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Amyloid beta A4 protein, GANTENERUMAB FAB FRAGMENT HEAVY CHAIN, ... | | 著者 | Benz, J, Burger, D, Loetscher, H.R, Bohrmann, B. | | 登録日 | 2015-07-23 | | 公開日 | 2015-08-12 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Gantenerumab: a novel human anti-Abeta antibody demonstrates sustained cerebral amyloid-Beta binding and elicits cell-mediated removal of human amyloid-Beta.
J. Alzheimers Dis., 28, 2012
|
|
1BGZ
 
 | |
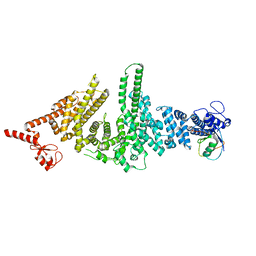
3M63
 
 | |
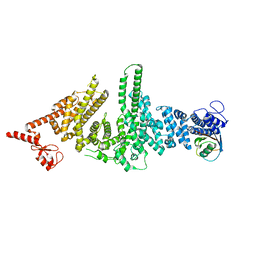
3M62
 
 | |
6FSM
 
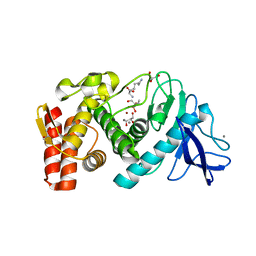 | | Crystal structure of TCE-treated Thermolysin | | 分子名称: | CALCIUM ION, GLYCEROL, LYSINE, ... | | 著者 | Pichlo, C, Baumann, U. | | 登録日 | 2018-02-19 | | 公開日 | 2018-05-02 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.39 Å) | | 主引用文献 | Improved protein-crystal identification by using 2,2,2-trichloroethanol as a fluorescence enhancer.
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
3FGW
 
 | | One chain form of the 66.3 kDa protein | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, IODIDE ION, ... | | 著者 | Lakomek, K, Dickmanns, A, Ficner, R. | | 登録日 | 2008-12-08 | | 公開日 | 2009-09-15 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Initial insight into the function of the lysosomal 66.3 kDa protein from mouse by means of X-ray crystallography
Bmc Struct.Biol., 9, 2009
|
|
6FSJ
 
 | | Crystal structure of TCE-treated Lysozyme | | 分子名称: | 1,2-ETHANEDIOL, CITRIC ACID, Lysozyme C | | 著者 | Pichlo, C, Baumann, U. | | 登録日 | 2018-02-19 | | 公開日 | 2018-05-02 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.2 Å) | | 主引用文献 | Improved protein-crystal identification by using 2,2,2-trichloroethanol as a fluorescence enhancer.
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
7LJR
 
 | | SARS-CoV-2 Spike Protein Trimer bound to DH1043 fab | | 分子名称: | Fab DH1043 heavy chain, Fab DH1043 light chain, Spike glycoprotein | | 著者 | Gobeil, S, Acharya, P. | | 登録日 | 2021-01-30 | | 公開日 | 2021-03-31 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (3.66 Å) | | 主引用文献 | The functions of SARS-CoV-2 neutralizing and infection-enhancing antibodies in vitro and in mice and nonhuman primates.
Biorxiv, 2021
|
|
3FGT
 
 | | Two chain form of the 66.3 kDa protein from mouse lacking the linker peptide | | 分子名称: | 2-(2-(2-(2-(2-(2-ETHOXYETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHANOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Lakomek, K, Dickmanns, A, Ficner, R. | | 登録日 | 2008-12-08 | | 公開日 | 2009-09-15 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Initial insight into the function of the lysosomal 66.3 kDa protein from mouse by means of X-ray crystallography
Bmc Struct.Biol., 9, 2009
|
|
3FGR
 
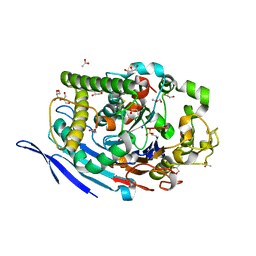 | | Two chain form of the 66.3 kDa protein at 1.8 Angstroem | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, ... | | 著者 | Lakomek, K, Dickmanns, A, Ficner, R. | | 登録日 | 2008-12-08 | | 公開日 | 2009-09-15 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Initial insight into the function of the lysosomal 66.3 kDa protein from mouse by means of X-ray crystallography
Bmc Struct.Biol., 9, 2009
|
|
7M0J
 
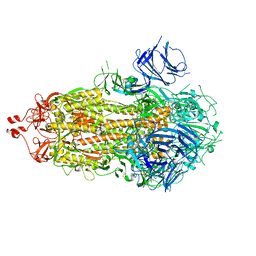 | |
7LWS
 
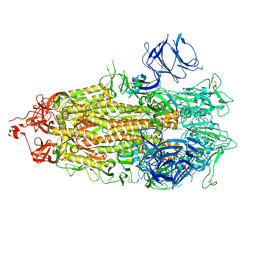 | |
8DKF
 
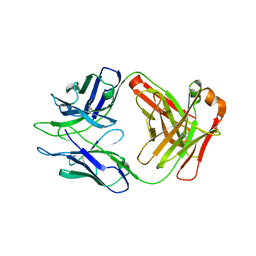 | |
3LQ2
 
 | | E. coli pyruvate dehydrogenase complex E1 E235A mutant with low TDP concentration | | 分子名称: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, MAGNESIUM ION, PHOSPHATE ION, ... | | 著者 | Furey, W. | | 登録日 | 2010-02-08 | | 公開日 | 2010-03-02 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.96 Å) | | 主引用文献 | Communication between thiamin cofactors in the Escherichia coli pyruvate dehydrogenase complex E1 component active centers: evidence for a "direct pathway" between the 4'-aminopyrimidine N1' atoms.
J.Biol.Chem., 285, 2010
|
|
3LQ4
 
 | | E. coli pyruvate dehydrogenase complex E1 E235A mutant with high TDP concentration | | 分子名称: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, MAGNESIUM ION, PHOSPHATE ION, ... | | 著者 | Furey, W. | | 登録日 | 2010-02-08 | | 公開日 | 2010-03-02 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.98 Å) | | 主引用文献 | Communication between thiamin cofactors in the Escherichia coli pyruvate dehydrogenase complex E1 component active centers: evidence for a "direct pathway" between the 4'-aminopyrimidine N1' atoms.
J.Biol.Chem., 285, 2010
|
|
3LPL
 
 | | E. coli pyruvate dehydrogenase complex E1 component E571A mutant | | 分子名称: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, MAGNESIUM ION, PHOSPHATE ION, ... | | 著者 | Furey, W. | | 登録日 | 2010-02-05 | | 公開日 | 2010-03-02 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Communication between thiamin cofactors in the Escherichia coli pyruvate dehydrogenase complex E1 component active centers: evidence for a "direct pathway" between the 4'-aminopyrimidine N1' atoms.
J.Biol.Chem., 285, 2010
|
|
