8HVM
 
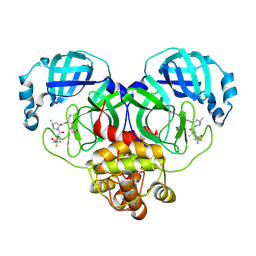 | | Crystal structure of SARS-Cov-2 main protease K90R mutant in complex with PF07321332 | | 分子名称: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | 著者 | Wang, J, Zhang, J, Li, J. | | 登録日 | 2022-12-27 | | 公開日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.48 Å) | | 主引用文献 | Crystal structure of SARS-Cov-2 main protease
K90R mutant in complex with PF07321332
To Be Published
|
|
8HVY
 
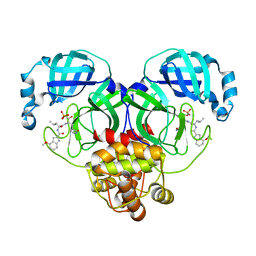 | |
6LAN
 
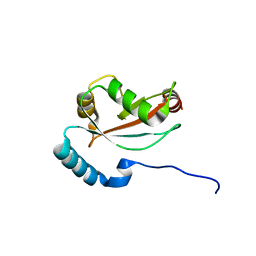 | | Structure of CCDC50 and LC3B complex | | 分子名称: | Coiled-coil domain-containing protein 50,Microtubule-associated proteins 1A/1B light chain 3B | | 著者 | Liu, L, Li, J, Hou, P. | | 登録日 | 2019-11-12 | | 公開日 | 2020-09-30 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.41 Å) | | 主引用文献 | A novel selective autophagy receptor, CCDC50, delivers K63 polyubiquitination-activated RIG-I/MDA5 for degradation during viral infection.
Cell Res., 31, 2021
|
|
8HVU
 
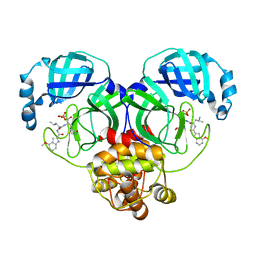 | |
8HVX
 
 | |
8HVW
 
 | |
8HVV
 
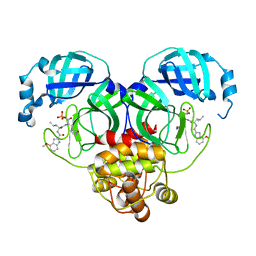 | |
8VJC
 
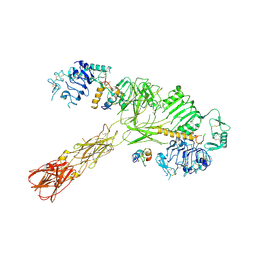 | | Cryo-EM structure of short form insulin receptor (IR-A) with three IGF2 bound, asymmetric conformation. | | 分子名称: | Insulin-like growth factor II, Isoform Short of Insulin receptor | | 著者 | An, W, Hall, C, Li, J, Huang, A, Wu, J, Park, J, Bai, X.C, Choi, E. | | 登録日 | 2024-01-06 | | 公開日 | 2024-03-27 | | 最終更新日 | 2024-04-03 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Activation of the insulin receptor by insulin-like growth factor 2.
Nat Commun, 15, 2024
|
|
8VJB
 
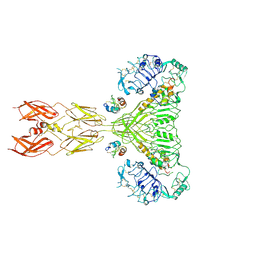 | | Cryo-EM structure of short form insulin receptor (IR-A) with four IGF2 bound, symmetric conformation. | | 分子名称: | Insulin-like growth factor II, Isoform Short of Insulin receptor | | 著者 | An, W, Hall, C, Li, J, Huang, A, Wu, J, Park, J, Bai, X.C, Choi, E. | | 登録日 | 2024-01-06 | | 公開日 | 2024-03-27 | | 最終更新日 | 2024-04-03 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Activation of the insulin receptor by insulin-like growth factor 2.
Nat Commun, 15, 2024
|
|
4WZJ
 
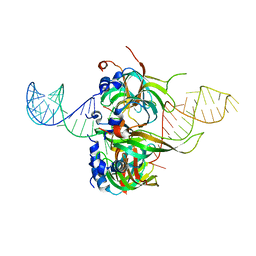 | | Spliceosomal U4 snRNP core domain | | 分子名称: | Small nuclear ribonucleoprotein E, Small nuclear ribonucleoprotein F, Small nuclear ribonucleoprotein G, ... | | 著者 | Leung, A.K.W, Nagai, K, Li, J. | | 登録日 | 2014-11-19 | | 公開日 | 2015-01-14 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (3.6 Å) | | 主引用文献 | Structure of the spliceosomal U4 snRNP core domain and its implication for snRNP biogenesis.
Nature, 473, 2011
|
|
8WZQ
 
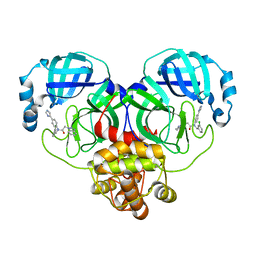 | | Crystal structure of SARS-Cov-2 main protease V186F mutant in complex with CCF0058981 | | 分子名称: | 2-(benzotriazol-1-yl)-~{N}-[(3-chlorophenyl)methyl]-~{N}-[4-(1~{H}-imidazol-5-yl)phenyl]ethanamide, 3C-like proteinase nsp5 | | 著者 | Zou, X.F, Jiang, H.H, Zhou, X.L, Zhang, J, Li, J. | | 登録日 | 2023-11-02 | | 公開日 | 2024-04-17 | | 実験手法 | X-RAY DIFFRACTION (1.66 Å) | | 主引用文献 | Crystal structure of SARS-CoV-2 main protease (M pro ) mutants in complex with the non-covalent inhibitor CCF0058981.
Biochem.Biophys.Res.Commun., 692, 2024
|
|
8WZP
 
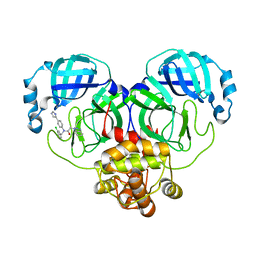 | | Crystal structure of SARS-Cov-2 main protease M49I mutant in complex with CCF0058981 | | 分子名称: | 2-(benzotriazol-1-yl)-~{N}-[(3-chlorophenyl)methyl]-~{N}-[4-(1~{H}-imidazol-5-yl)phenyl]ethanamide, 3C-like proteinase nsp5 | | 著者 | Jiang, H.H, Zou, X.F, Zhou, X.L, Zhang, J, Li, J. | | 登録日 | 2023-11-02 | | 公開日 | 2024-04-17 | | 実験手法 | X-RAY DIFFRACTION (1.76 Å) | | 主引用文献 | Crystal structure of SARS-CoV-2 main protease (M pro ) mutants in complex with the non-covalent inhibitor CCF0058981.
Biochem.Biophys.Res.Commun., 692, 2024
|
|
6LPF
 
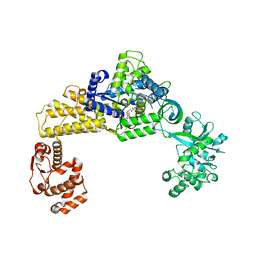 | | The crystal structure of human cytoplasmic LRS | | 分子名称: | 2'-(L-NORVALYL)AMINO-2'-DEOXYADENOSINE, 5'-O-(L-leucylsulfamoyl)adenosine, GLYCEROL, ... | | 著者 | Liu, R.J, Long, T, Li, H, Li, J, Zhao, J.H, Lin, J.Z, Palencia, A, Wang, M.Z, Cusack, S, Wang, E.D. | | 登録日 | 2020-01-10 | | 公開日 | 2020-03-25 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.49 Å) | | 主引用文献 | Molecular basis of the multifaceted functions of human leucyl-tRNA synthetase in protein synthesis and beyond.
Nucleic Acids Res., 48, 2020
|
|
8H91
 
 | |
6KOB
 
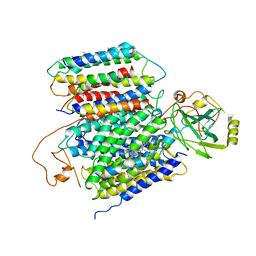 | | X-ray Structure of the proton-pumping cytochrome aa3-600 menaquinol oxidase from Bacillus subtilis | | 分子名称: | AA3-600 quinol oxidase subunit I, AA3-600 quinol oxidase subunit IIII, AA3-600 quinol oxidase subunit IV,Quinol oxidase subunit 4, ... | | 著者 | Xu, J, Ding, Z, Liu, B, Li, J, Gennis, R.B, Zhu, J. | | 登録日 | 2019-08-09 | | 公開日 | 2020-01-15 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (3.6 Å) | | 主引用文献 | Structure of the cytochromeaa3-600 heme-copper menaquinol oxidase bound to inhibitor HQNO shows TM0 is part of the quinol binding site.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
8HQJ
 
 | |
8HQH
 
 | |
8HVK
 
 | | Crystal structure of SARS-Cov-2 main protease G15S mutant in complex with PF07321332 | | 分子名称: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | 著者 | Zeng, P, Zhang, J, Li, J. | | 登録日 | 2022-12-27 | | 公開日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.63 Å) | | 主引用文献 | Crystal structure of SARS-Cov-2 main protease
G15S mutant in complex with PF07321332
To Be Published
|
|
8HVO
 
 | | Crystal structure of SARS-Cov-2 main protease V186F mutant in complex with PF07321332 | | 分子名称: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | 著者 | Zou, X.F, Zhang, J, Li, J. | | 登録日 | 2022-12-27 | | 公開日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Crystal structure of SARS-Cov-2 main protease
V186F mutant in complex with PF07321332
To Be Published
|
|
8HVL
 
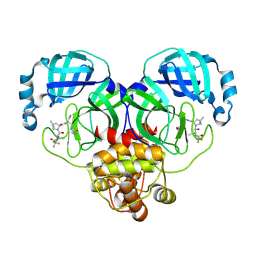 | | Crystal structure of SARS-Cov-2 main protease M49I mutant in complex with PF07321332 | | 分子名称: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | 著者 | Zou, X.F, Zhang, J, Li, J. | | 登録日 | 2022-12-27 | | 公開日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | Crystal structure of SARS-Cov-2 main protease
M49I mutant in complex with PF07321332
To Be Published
|
|
8HQF
 
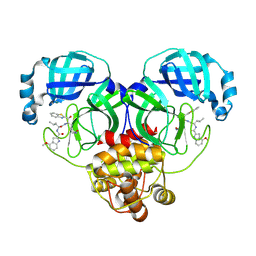 | |
3L3Z
 
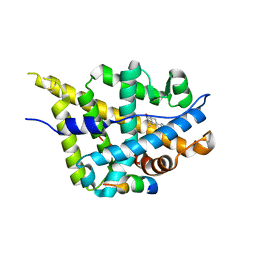 | | Crystal structure of DHT-bound androgen receptor in complex with the third motif of steroid receptor coactivator 3 | | 分子名称: | 5-ALPHA-DIHYDROTESTOSTERONE, Androgen receptor, Nuclear receptor coactivator 3 | | 著者 | Zhou, X.E, Suino-Powell, K.M, Li, J, He, A, MacKeigan, J.P, Melcher, K, Yong, E.-L, Xu, H.E. | | 登録日 | 2009-12-18 | | 公開日 | 2010-01-12 | | 最終更新日 | 2017-11-01 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Identification of SRC3/AIB1 as a Preferred Coactivator for Hormone-activated Androgen Receptor.
J.Biol.Chem., 285, 2010
|
|
3L3X
 
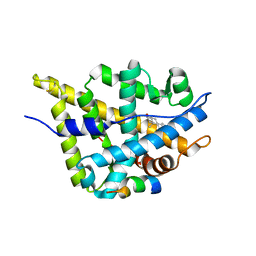 | | Crystal structure of DHT-bound androgen receptor in complex with the first motif of steroid receptor coactivator 3 | | 分子名称: | 5-ALPHA-DIHYDROTESTOSTERONE, Androgen receptor, Nuclear receptor coactivator 3 | | 著者 | Zhou, X.E, Suino-Powell, K.M, Li, J, He, A, MacKeigan, J.P, Melcher, K, Yong, E.-L, Xu, H.E. | | 登録日 | 2009-12-18 | | 公開日 | 2010-01-12 | | 最終更新日 | 2017-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Identification of SRC3/AIB1 as a Preferred Coactivator for Hormone-activated Androgen Receptor.
J.Biol.Chem., 285, 2010
|
|
8YKW
 
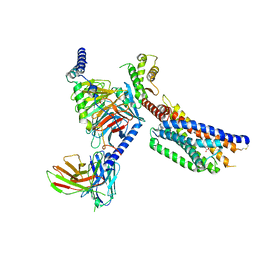 | | Cryo-EM structure of succinate receptor SUCR1 bound to succinic acid | | 分子名称: | Antibody fragment ScFv16, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | 著者 | Li, C, Liu, H, Li, J, Zhu, H, Fu, W, Xu, H.E. | | 登録日 | 2024-03-05 | | 公開日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (2.75 Å) | | 主引用文献 | Cryo-EM structure of succinate receptor SUCR1 bound to succinic acid
To Be Published
|
|
8YKV
 
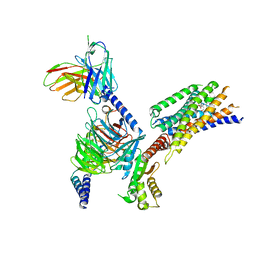 | | Cryo-EM structure of succinate receptor SUCR1 bound to compound 31 | | 分子名称: | (2~{S})-2-[[6-[4-(trifluoromethyloxy)phenyl]pyridin-2-yl]carbonylamino]butanedioic acid, Antibody fragment ScFv16, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | 著者 | Li, C, Liu, H, Li, J, Zhu, H, Fu, W, Xu, H.E. | | 登録日 | 2024-03-05 | | 公開日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (2.48 Å) | | 主引用文献 | Cryo-EM structure of succinate receptor SUCR1 bound to compound 31
To Be Published
|
|
