6DKM
 
 | | DHD131 | | 分子名称: | DHD131_A, DHD131_B | | 著者 | Bick, M.J, Chen, Z, Baker, D. | | 登録日 | 2018-05-29 | | 公開日 | 2018-12-19 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.38 Å) | | 主引用文献 | Programmable design of orthogonal protein heterodimers.
Nature, 565, 2019
|
|
6DM9
 
 | | DHD15_extended | | 分子名称: | DHD15_extended_A, DHD15_extended_B, SULFATE ION | | 著者 | Bick, M.J, Chen, Z, Baker, D. | | 登録日 | 2018-06-04 | | 公開日 | 2018-12-19 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Programmable design of orthogonal protein heterodimers.
Nature, 565, 2019
|
|
6DLC
 
 | |
6C2V
 
 | | Solution structure of a phosphate-loop protein | | 分子名称: | phosphate-loop protein | | 著者 | Yang, F, Yang, W, Lin, Y.R, Romero Romero, M.L, Tawfik, D, Baker, D, Varani, G. | | 登録日 | 2018-01-09 | | 公開日 | 2018-11-21 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Simple yet functional phosphate-loop proteins.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6C2U
 
 | | Solution structure of a phosphate-loop protein | | 分子名称: | phosphate-loop protein | | 著者 | Yang, F, Yang, W, Lin, Y.R, Romero Romero, M.L, Tawfik, D, Baker, D, Varani, G. | | 登録日 | 2018-01-09 | | 公開日 | 2018-11-21 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Simple yet functional phosphate-loop proteins.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5W9F
 
 | | Solution structure of the de novo mini protein gHEEE_02 | | 分子名称: | De novo mini protein gHEEE_02 | | 著者 | Pulavarti, S.V.S.R.K, Shaw, E.A, Bahl, C.D, Garry, B.W, Baker, D, Szyperski, T. | | 登録日 | 2017-06-23 | | 公開日 | 2018-07-11 | | 最終更新日 | 2023-06-14 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Cytosolic expression, solution structures, and molecular dynamics simulation of genetically encodable disulfide-rich de novo designed peptides.
Protein Sci., 27, 2018
|
|
7KUW
 
 | |
7UCP
 
 | | computationally designed macrocycle | | 分子名称: | computationally designed cyclic peptide D8.3.p2 | | 著者 | Bhardwaj, G, Baker, D, Rettie, S, Glynn, C, Sawaya, M. | | 登録日 | 2022-03-17 | | 公開日 | 2022-09-14 | | 最終更新日 | 2022-09-28 | | 実験手法 | X-RAY DIFFRACTION (0.85 Å) | | 主引用文献 | Accurate de novo design of membrane-traversing macrocycles.
Cell, 185, 2022
|
|
4A29
 
 | | Structure of the engineered retro-aldolase RA95.0 | | 分子名称: | 1-(6-METHOXYNAPHTHALEN-2-YL)BUTANE-1,3-DIONE, D-MALATE, ENGINEERED RETRO-ALDOL ENZYME RA95.0 | | 著者 | Giger, L, Caner, S, Kast, P, Baker, D, Ban, N, Hilvert, D. | | 登録日 | 2011-09-23 | | 公開日 | 2012-11-07 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.1 Å) | | 主引用文献 | Evolution of a designed retro-aldolase leads to complete active site remodeling.
Nat.Chem.Biol., 9, 2013
|
|
4A2R
 
 | | Structure of the engineered retro-aldolase RA95.5-5 | | 分子名称: | 1-(6-METHOXYNAPHTHALEN-2-YL)BUTANE-1,3-DIONE, INDOLE-3-GLYCEROL PHOSPHATE SYNTHASE | | 著者 | Giger, L, Caner, S, Kast, P, Baker, D, Ban, N, Hilvert, D. | | 登録日 | 2011-09-28 | | 公開日 | 2012-11-07 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.302 Å) | | 主引用文献 | Evolution of a designed retro-aldolase leads to complete active site remodeling.
Nat.Chem.Biol., 9, 2013
|
|
4A2S
 
 | | Structure of the engineered retro-aldolase RA95.5 | | 分子名称: | 1-(6-METHOXYNAPHTHALEN-2-YL)BUTANE-1,3-DIONE, INDOLE-3-GLYCEROL PHOSPHATE SYNTHASE | | 著者 | Giger, L, Caner, S, Kast, P, Baker, D, Ban, N, Hilvert, D. | | 登録日 | 2011-09-28 | | 公開日 | 2012-11-07 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Evolution of a designed retro-aldolase leads to complete active site remodeling.
Nat.Chem.Biol., 9, 2013
|
|
2FLD
 
 | | I-MsoI Re-Designed for Altered DNA Cleavage Specificity | | 分子名称: | 5'-D(*CP*GP*GP*AP*AP*CP*GP*GP*TP*CP*TP*CP*AP*CP*GP*AP*CP*CP*TP*TP*CP*TP*GP*C)-3', 5'-D(*GP*CP*AP*GP*AP*AP*GP*GP*TP*CP*GP*TP*GP*AP*GP*AP*CP*CP*GP*TP*TP*CP*CP*G)-3', CALCIUM ION, ... | | 著者 | Ashworth, J, Duarte, C.M, Havranek, J.J, Sussman, D, Monnat, R.J, Stoddard, B.L, Baker, D. | | 登録日 | 2006-01-05 | | 公開日 | 2006-06-06 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Computational redesign of endonuclease DNA binding and cleavage specificity.
Nature, 441, 2006
|
|
4CDL
 
 | | Crystal Structure of Retro-aldolase RA110.4-6 Complexed with Inhibitor 1-(6-methoxy-2-naphthalenyl)-1,3-butanedione | | 分子名称: | (2E)-1-(6-methoxynaphthalen-2-yl)but-2-en-1-one, STEROID DELTA-ISOMERASE | | 著者 | Pinkas, D.M, Studer, S, Obexer, R, Giger, L, Gruetter, M.G, Baker, D, Hilvert, D. | | 登録日 | 2013-11-01 | | 公開日 | 2014-11-12 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Active Site Plasticity of a Computationally Designed Retro-Aldolase Enzyme
To be Published
|
|
5EC0
 
 | | Crystal Structure of Actin-like protein Alp7A | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Alp7A, GLYCEROL, ... | | 著者 | Petek, N.A, Kraemer, J.A, Mullins, R.D, Agard, D.A, DiMaio, F, Baker, D. | | 登録日 | 2015-10-20 | | 公開日 | 2016-11-02 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Structure and function of Alp7A reveal both conserved and unique features of plasmid segregation.
To Be Published
|
|
5TVY
 
 | | Computationally Designed Fentanyl Binder - Fen49 | | 分子名称: | 3,6,9,12,15,18,21,24,27-NONAOXANONACOSANE-1,29-DIOL, Endo-1,4-beta-xylanase A | | 著者 | Bick, M.J, Greisen, P.J, Morey, K.J, Antunes, M.S, La, D, Sankaran, B, Reymond, L, Johnsson, K, Medford, J.I, Baker, D. | | 登録日 | 2016-11-10 | | 公開日 | 2017-10-04 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1 Å) | | 主引用文献 | Computational design of environmental sensors for the potent opioid fentanyl.
Elife, 6, 2017
|
|
3GEQ
 
 | | Structural basis for the chemical rescue of Src kinase activity | | 分子名称: | 1-TERT-BUTYL-3-(4-CHLORO-PHENYL)-1H-PYRAZOLO[3,4-D]PYRIMIDIN-4-YLAMINE, Proto-oncogene tyrosine-protein kinase Src | | 著者 | Muratore, K.E, Seeliger, M.A, Wang, Z, Fomina, D, Neiswinger, J, Havranek, J.J, Baker, D, Kuriyan, J, Cole, P.A. | | 登録日 | 2009-02-25 | | 公開日 | 2009-04-28 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Comparative analysis of mutant tyrosine kinase chemical rescue.
Biochemistry, 48, 2009
|
|
7CBC
 
 | |
5TVV
 
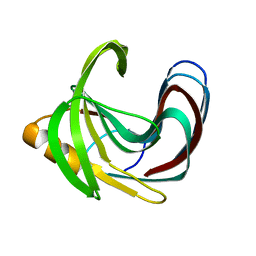 | | Computationally Designed Fentanyl Binder - Fen49* Apo | | 分子名称: | Endo-1,4-beta-xylanase A, POTASSIUM ION | | 著者 | Bick, M.J, Greisen, P.J, Morey, K.J, Antunes, A.S, La, D, Sankaran, B, Reymond, L, Johnsson, K, Medford, J.I, Baker, D. | | 登録日 | 2016-11-10 | | 公開日 | 2017-10-04 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.79 Å) | | 主引用文献 | Computational design of environmental sensors for the potent opioid fentanyl.
Elife, 6, 2017
|
|
5EIL
 
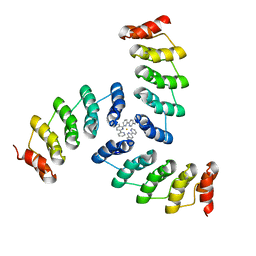 | | Computational design of a high-affinity metalloprotein homotrimer containing a metal chelating non-canonical amino acid | | 分子名称: | FE (III) ION, TRI-05 | | 著者 | Sankaran, B, Zwart, P.H, Mills, J.H, Pereira, J.H, Baker, D. | | 登録日 | 2015-10-30 | | 公開日 | 2016-11-16 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Computational design of a homotrimeric metalloprotein with a trisbipyridyl core.
Proc. Natl. Acad. Sci. U.S.A., 113, 2016
|
|
5TZO
 
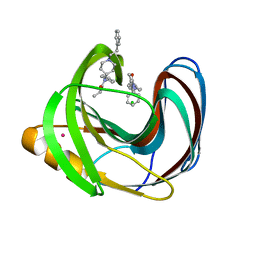 | | Computationally Designed Fentanyl Binder - Fen49*-Complex | | 分子名称: | CHLORIDE ION, Endo-1,4-beta-xylanase A, N-phenyl-N-[1-(2-phenylethyl)piperidin-4-yl]propanamide, ... | | 著者 | Bick, M.J, Greisen, P.J, Morey, K.J, Antunes, M.S, La, D, Sankaran, B, Reymond, L, Johnsson, K, Medford, J.I, Baker, D. | | 登録日 | 2016-11-22 | | 公開日 | 2017-10-04 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.67 Å) | | 主引用文献 | Computational design of environmental sensors for the potent opioid fentanyl.
Elife, 6, 2017
|
|
8FRE
 
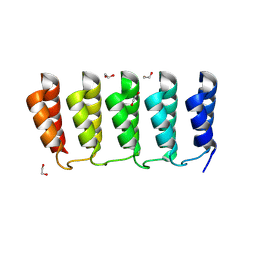 | | Designed loop protein RBL4 | | 分子名称: | 1,2-ETHANEDIOL, RBL4 | | 著者 | Jude, K.M, Jiang, H, Baker, D, Garcia, K.C. | | 登録日 | 2023-01-06 | | 公開日 | 2024-04-17 | | 最終更新日 | 2024-06-26 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | De novo design of buttressed loops for sculpting protein functions.
Nat.Chem.Biol., 2024
|
|
8FRF
 
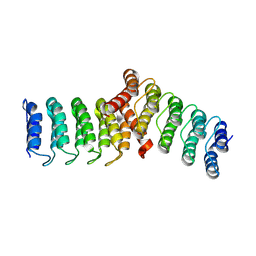 | |
8FNE
 
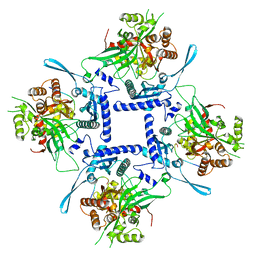 | | phiPA3 PhuN Tetramer, p2 | | 分子名称: | Maltose/maltodextrin-binding periplasmic protein, PhuN | | 著者 | Nieweglowska, E.S, Brilot, A.F, Mendez-Moran, M, Kokontis, C, Baek, M, Li, J, Cheng, Y, Baker, D, Bondy-Denomy, J, Agard, D.A. | | 登録日 | 2022-12-27 | | 公開日 | 2023-03-01 | | 最終更新日 | 2024-06-19 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | The phi PA3 phage nucleus is enclosed by a self-assembling 2D crystalline lattice.
Nat Commun, 14, 2023
|
|
8FV5
 
 | | Representation of 16-mer phiPA3 PhuN Lattice, p2 | | 分子名称: | Maltose/maltodextrin-binding periplasmic protein, phiPA3 PhuN | | 著者 | Nieweglowska, E.S, Brilot, A.F, Mendez-Moran, M, Kokontis, C, Baek, M, Li, J, Cheng, Y, Baker, D, Bondy-Denomy, J, Agard, D.A. | | 登録日 | 2023-01-18 | | 公開日 | 2023-03-01 | | 最終更新日 | 2024-05-01 | | 実験手法 | ELECTRON MICROSCOPY (4.21 Å) | | 主引用文献 | The phi PA3 phage nucleus is enclosed by a self-assembling 2D crystalline lattice.
Nat Commun, 14, 2023
|
|
1VJQ
 
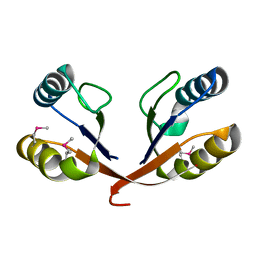 | |
