7ECW
 
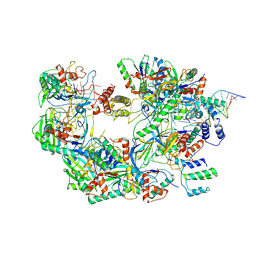 | | The Csy-AcrIF14-dsDNA complex | | 分子名称: | 54-MER DNA, AcrIF14, CRISPR type I-F/YPEST-associated protein Csy2, ... | | 著者 | Zhang, L.X, Feng, Y. | | 登録日 | 2021-03-13 | | 公開日 | 2021-11-17 | | 最終更新日 | 2024-06-05 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Insights into the dual functions of AcrIF14 during the inhibition of type I-F CRISPR-Cas surveillance complex.
Nucleic Acids Res., 49, 2021
|
|
7WHI
 
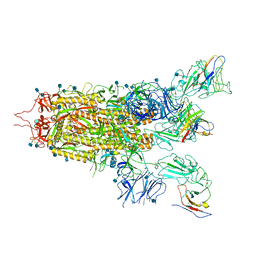 | | The state 2 complex structure of Omicron spike with Bn03 (2-up RBD, 4 nanobodies) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Bn03_nano1, Bn03_nano2, ... | | 著者 | Zhan, W.Q, Zhang, X, Chen, Z.G, Sun, L. | | 登録日 | 2021-12-30 | | 公開日 | 2022-05-11 | | 実験手法 | ELECTRON MICROSCOPY (2.93 Å) | | 主引用文献 | Broad neutralization of SARS-CoV-2 variants by an inhalable bispecific single-domain antibody.
Cell, 185, 2022
|
|
7WHJ
 
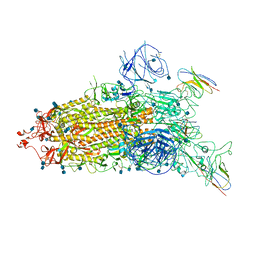 | | The state 1 complex structure of Omicron spike with Bn03 (1-up RBD, 3 nanobodies) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Bn03_nano1, Bn03_nano2, ... | | 著者 | Zhan, W.Q, Zhang, X, Chen, Z.G, Sun, L. | | 登録日 | 2021-12-30 | | 公開日 | 2022-05-11 | | 実験手法 | ELECTRON MICROSCOPY (3.27 Å) | | 主引用文献 | Broad neutralization of SARS-CoV-2 variants by an inhalable bispecific single-domain antibody.
Cell, 185, 2022
|
|
7WHK
 
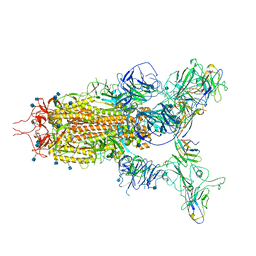 | | The state 3 complex structure of Omicron spike with Bn03 (2-up RBD, 5 nanobodies) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Bn03_nano1, Bn03_nano2, ... | | 著者 | Zhan, W.Q, Zhang, X, Chen, Z.G, Sun, L. | | 登録日 | 2021-12-30 | | 公開日 | 2022-05-11 | | 実験手法 | ELECTRON MICROSCOPY (3.01 Å) | | 主引用文献 | Broad neutralization of SARS-CoV-2 variants by an inhalable bispecific single-domain antibody.
Cell, 185, 2022
|
|
7X9E
 
 | | Crystal structure of the 76E1 Fab in complex with a SARS-CoV-2 spike peptide | | 分子名称: | 76E1 Fab Heavy Chain, 76E1 Fab Light Chain, Spike peptide | | 著者 | Chen, X, Zhang, T, Ding, J, Sun, X, Sun, B. | | 登録日 | 2022-03-15 | | 公開日 | 2022-05-11 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Neutralization mechanism of a human antibody with pan-coronavirus reactivity including SARS-CoV-2.
Nat Microbiol, 7, 2022
|
|
7Y53
 
 | | The cryo-EM structure of human ERAD retro-translocation complex | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Derlin-1, Transitional endoplasmic reticulum ATPase | | 著者 | Cao, Y, Rao, B, Wang, Q, Yao, D, Xia, Y, Li, W, Li, S, Shen, Y. | | 登録日 | 2022-06-16 | | 公開日 | 2023-10-18 | | 最終更新日 | 2024-04-03 | | 実験手法 | ELECTRON MICROSCOPY (3.61 Å) | | 主引用文献 | The cryo-EM structure of the human ERAD retrotranslocation complex.
Sci Adv, 9, 2023
|
|
7Y4W
 
 | | The cryo-EM structure of human ERAD retro-translocation complex | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Derlin-1, Transitional endoplasmic reticulum ATPase | | 著者 | Cao, Y, Rao, B, Wang, Q, Yao, D, Xia, Y, Li, W, Li, S, Shen, Y. | | 登録日 | 2022-06-16 | | 公開日 | 2023-10-18 | | 最終更新日 | 2024-04-03 | | 実験手法 | ELECTRON MICROSCOPY (3.67 Å) | | 主引用文献 | The cryo-EM structure of the human ERAD retrotranslocation complex.
Sci Adv, 9, 2023
|
|
7Y59
 
 | | The cryo-EM structure of human ERAD retro-translocation complex | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Derlin-1, ... | | 著者 | Cao, Y, Rao, B, Wang, Q, Yao, D, Xia, Y, Li, W, Li, S, Shen, Y. | | 登録日 | 2022-06-16 | | 公開日 | 2023-10-18 | | 最終更新日 | 2024-04-03 | | 実験手法 | ELECTRON MICROSCOPY (4.51 Å) | | 主引用文献 | The cryo-EM structure of the human ERAD retrotranslocation complex.
Sci Adv, 9, 2023
|
|
7XCP
 
 | | Cryo-EM structure of Omicron RBD complexed with ACE2 and 304 Fab | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of 304 Fab, ... | | 著者 | Zhao, Z, Qi, J, Gao, F.G. | | 登録日 | 2022-03-24 | | 公開日 | 2022-08-31 | | 実験手法 | ELECTRON MICROSCOPY (3.05 Å) | | 主引用文献 | Omicron SARS-CoV-2 mutations stabilize spike up-RBD conformation and lead to a non-RBM-binding monoclonal antibody escape
Nat Commun, 13, 2022
|
|
7Y9Z
 
 | | Cryo-EM structure of SARS-CoV-2 Omicron spike protein (S-6P-RRAR) in complex with human ACE2 ectodomain (one-RBD-up state) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | 著者 | Gao, G.F, Qi, J.X, Liu, S, Zhao, Z.N. | | 登録日 | 2022-06-26 | | 公開日 | 2022-09-21 | | 実験手法 | ELECTRON MICROSCOPY (2.85 Å) | | 主引用文献 | Omicron SARS-CoV-2 mutations stabilize spike up-RBD conformation and lead to a non-RBM-binding monoclonal antibody escape.
Nat Commun, 13, 2022
|
|
7CT5
 
 | | S protein of SARS-CoV-2 in complex bound with T-ACE2 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | 著者 | Guo, L, Bi, W.W, Zhang, Y.Y, Yan, R.H, Li, Y.N, Zhou, Q, Dang, B.B. | | 登録日 | 2020-08-18 | | 公開日 | 2020-11-18 | | 最終更新日 | 2021-02-10 | | 実験手法 | ELECTRON MICROSCOPY (4 Å) | | 主引用文献 | Engineered trimeric ACE2 binds viral spike protein and locks it in "Three-up" conformation to potently inhibit SARS-CoV-2 infection.
Cell Res., 31, 2021
|
|
7DFH
 
 | |
7DFG
 
 | |
7DOI
 
 | |
7DOK
 
 | |
7XN2
 
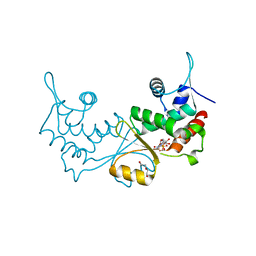 | | Crystal structure of CvkR, a novel MerR-type transcriptional regulator | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, Alr3614 protein, DI(HYDROXYETHYL)ETHER | | 著者 | Liang, Y.J, Zhu, T, Ma, H.L, Lu, X.F, Hess, W.R. | | 登録日 | 2022-04-27 | | 公開日 | 2023-03-08 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | CvkR is a MerR-type transcriptional repressor of class 2 type V-K CRISPR-associated transposase systems.
Nat Commun, 14, 2023
|
|
7VQ7
 
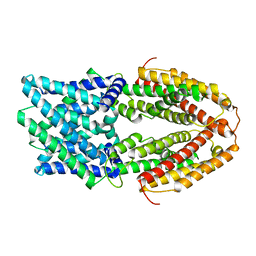 | |
7VQ5
 
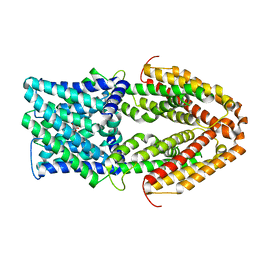 | |
7VQ3
 
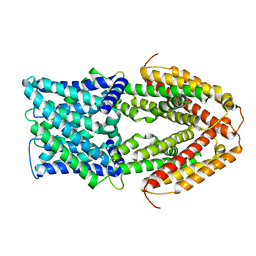 | |
7VQ4
 
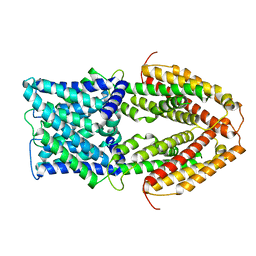 | |
7VOJ
 
 | | Al-bound structure of the AtALMT1 mutant M60A | | 分子名称: | ACETIC ACID, ALUMINUM ION, Aluminum-activated malate transporter 1 | | 著者 | Wang, J. | | 登録日 | 2021-10-14 | | 公開日 | 2021-12-01 | | 最終更新日 | 2022-02-16 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Structural basis of ALMT1-mediated aluminum resistance in Arabidopsis.
Cell Res., 32, 2022
|
|
7Y7J
 
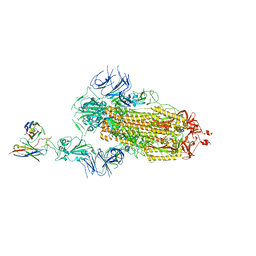 | | SARS-CoV-2 S trimer in complex with 1F Fab | | 分子名称: | 1F VH, 1F VL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Zhao, S, Liu, F, Yang, X, Zhong, G. | | 登録日 | 2022-06-22 | | 公開日 | 2023-02-22 | | 実験手法 | ELECTRON MICROSCOPY (4.8 Å) | | 主引用文献 | A core epitope targeting antibody of SARS-CoV-2.
Protein Cell, 14, 2023
|
|
7Y7K
 
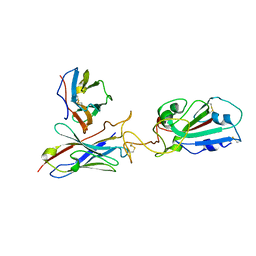 | | SARS-CoV-2 RBD in complex with 1F Fab | | 分子名称: | 1F VH, 1F VL, Spike protein S1 | | 著者 | Zhao, S, Liu, F, Yang, X, Zhong, G. | | 登録日 | 2022-06-22 | | 公開日 | 2023-02-22 | | 実験手法 | ELECTRON MICROSCOPY (4.4 Å) | | 主引用文献 | A core epitope targeting antibody of SARS-CoV-2.
Protein Cell, 14, 2023
|
|
7D2S
 
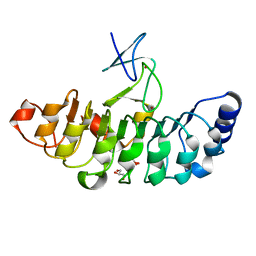 | | Crystal structure of Rsu1/PINCH1_LIM5C complex | | 分子名称: | GLYCEROL, LIM and senescent cell antigen-like-containing domain protein 1, Ras suppressor protein 1, ... | | 著者 | Yang, H, Wei, Z, Cong, Y. | | 登録日 | 2020-09-17 | | 公開日 | 2021-02-24 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.653 Å) | | 主引用文献 | Complex structures of Rsu1 and PINCH1 reveal a regulatory mechanism of the ILK/PINCH/Parvin complex for F-actin dynamics.
Elife, 10, 2021
|
|
7D2U
 
 | | Crystal structure of Rsu1/PINCH1_LIM45C complex | | 分子名称: | GLYCEROL, LIM and senescent cell antigen-like-containing domain protein 1, MALONATE ION, ... | | 著者 | Yang, H, Wei, Z, Cong, Y. | | 登録日 | 2020-09-17 | | 公開日 | 2021-02-24 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (3.15 Å) | | 主引用文献 | Complex structures of Rsu1 and PINCH1 reveal a regulatory mechanism of the ILK/PINCH/Parvin complex for F-actin dynamics.
Elife, 10, 2021
|
|
