2NP3
 
 | | Crystal structure of TetR-family regulator (SCO0857) from Streptomyces coelicolor A3. | | 分子名称: | Putative TetR-family regulator | | 著者 | Koclega, K.D, Xu, X, Chruszcz, M, Gu, J, Cymborowski, M, Savchenko, A, Edwards, A, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2006-10-26 | | 公開日 | 2006-11-28 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | Crystal structure of TetR-family regulator (SCO0857) from Streptomyces coelicolor A3.
To be Published
|
|
4FCG
 
 | | Structure of the leucine-rich repeat domain of the type III effector XCV3220 (XopL) | | 分子名称: | 1,2-ETHANEDIOL, CHLORIDE ION, PHOSPHATE ION, ... | | 著者 | Singer, A.U, Xu, X, Cui, H, Zimmerman, M.D, Minor, W, Joachimiak, A, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2012-05-24 | | 公開日 | 2012-06-13 | | 最終更新日 | 2022-04-13 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structure of the leucine-rich repeat domain of the type III effector XCV3220 (XopL)
To be Published
|
|
2QMO
 
 | | Crystal structure of dethiobiotin synthetase (bioD) from Helicobacter pylori | | 分子名称: | CHLORIDE ION, Dethiobiotin synthetase | | 著者 | Chruszcz, M, Xu, X, Cuff, M, Cymborowski, M, Zheng, H, Savchenko, A, Edwards, A, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2007-07-16 | | 公開日 | 2007-07-31 | | 最終更新日 | 2022-04-13 | | 実験手法 | X-RAY DIFFRACTION (1.47 Å) | | 主引用文献 | Structural characterization of Helicobacter pylori dethiobiotin synthetase reveals differences between family members.
Febs J., 279, 2012
|
|
2NP5
 
 | | Crystal structure of a transcriptional regulator (RHA1_ro04179) from Rhodococcus sp. Rha1. | | 分子名称: | DODECYL-BETA-D-MALTOSIDE, ETHYL DIMETHYL AMMONIO PROPANE SULFONATE, Transcriptional regulator | | 著者 | Chruszcz, M, Evdokimova, E, Kagan, O, Cymborowski, M, Savchenko, A, Edwards, A, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2006-10-26 | | 公開日 | 2006-11-28 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal structure of a transcriptional regulator (RHA1_ro04179) from Rhodococcus sp. Rha1.
TO BE PUBLISHED
|
|
7JH3
 
 | | Crystal structure of 4-aminobutyrate aminotransferase PuuE from Escherichia coli in complex with PLP | | 分子名称: | 4-aminobutyrate aminotransferase PuuE, DI(HYDROXYETHYL)ETHER | | 著者 | Valleau, D, Evdokimova, E, Stogios, P.J, Di Leo, R, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2020-07-20 | | 公開日 | 2020-08-12 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.68 Å) | | 主引用文献 | Crystal structure of 4-aminobutyrate aminotransferase PuuE from Escherichia coli in complex with PLP
To Be Published
|
|
7KAG
 
 | | Crystal structure of the ubiquitin-like domain 1 (Ubl1) of Nsp3 from SARS-CoV-2 | | 分子名称: | 1,2-ETHANEDIOL, Non-structural protein 3, SULFATE ION | | 著者 | Stogios, P.J, Skarina, T, Chang, C, Kim, Y, Di Leo, R, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2020-09-30 | | 公開日 | 2020-10-14 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (3.21 Å) | | 主引用文献 | Crystal structure of the ubiquitin-like domain 1 (Ubl1) of Nsp3 from SARS-CoV-2
To Be Published
|
|
7JM1
 
 | | Crystal structure of aminoglycoside resistance enzyme ApmA, complex with acetyl-CoA | | 分子名称: | ACETYL COENZYME *A, Aminocyclitol acetyltransferase ApmA | | 著者 | Stogios, P.J, Evdokimova, E, Di Leo, R, Bordeleau, E, Wright, G.D, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2020-07-30 | | 公開日 | 2020-09-16 | | 実験手法 | X-RAY DIFFRACTION (2.31 Å) | | 主引用文献 | Crystal structure of aminoglycoside resistance enzyme ApmA, complex with acetyl-CoA
To Be Published
|
|
7JM2
 
 | | Crystal structure of aminoglycoside resistance enzyme ApmA, complex with apramycin | | 分子名称: | APRAMYCIN, Aminocyclitol acetyltransferase ApmA, CHLORIDE ION | | 著者 | Stogios, P.J, Evdokimova, E, Di Leo, R, Bordeleau, E, Wright, G.D, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2020-07-30 | | 公開日 | 2020-09-16 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Crystal structure of aminoglycoside resistance enzyme ApmA, complex with apramycin
To Be Published
|
|
7JM0
 
 | | Crystal structure of aminoglycoside resistance enzyme ApmA, apoenzyme | | 分子名称: | Aminocyclitol acetyltransferase ApmA, SULFATE ION | | 著者 | Stogios, P.J, Evdokimova, E, Di Leo, R, Bordeleau, E, Wright, G.D, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2020-07-30 | | 公開日 | 2020-09-16 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.08 Å) | | 主引用文献 | Crystal structure of aminoglycoside resistance enzyme ApmA, apoenzyme
To Be Published
|
|
7LGO
 
 | | Crystal structure of the nucleic acid binding domain (NAB) of Nsp3 from SARS-CoV-2 | | 分子名称: | Non-structural protein 3 | | 著者 | Stogios, P.J, Skarina, T, Di Leo, R, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2021-01-20 | | 公開日 | 2021-01-27 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.45 Å) | | 主引用文献 | Crystal structure of the nucleic acid binding domain (NAB) of Nsp3 from SARS-CoV-2
To Be Published
|
|
2PFS
 
 | | Crystal structure of universal stress protein from Nitrosomonas europaea | | 分子名称: | CHLORIDE ION, Universal stress protein | | 著者 | Chruszcz, M, Evdokimova, E, Cymborowski, M, Kagan, O, Savchenko, A, Edwards, A, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2007-04-05 | | 公開日 | 2007-05-08 | | 最終更新日 | 2022-04-13 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Structural and functional insight into the universal stress protein family.
Evol Appl, 6, 2013
|
|
7KZW
 
 | | Crystal structure of FTT_1639c from Francisella tularensis str. tularensis SCHU S4 | | 分子名称: | CHLORIDE ION, FTT_1639c | | 著者 | Stogios, P.J, Skarina, T, Osipiuk, J, Di Leo, R, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2020-12-10 | | 公開日 | 2020-12-30 | | 実験手法 | X-RAY DIFFRACTION (1.34 Å) | | 主引用文献 | Crystal structure of FTT_1639c from Francisella tularensis str. tularensis SCHU S4
To Be Published
|
|
2Q24
 
 | | Crystal structure of TetR transcriptional regulator SCO0520 from Streptomyces coelicolor | | 分子名称: | ACETATE ION, CHLORIDE ION, Putative tetR family transcriptional regulator | | 著者 | Cymborowski, M, Chruszcz, M, Koclega, K.D, Filippova, E.V, Xu, X, Gu, J, Savchenko, A, Edwards, A.M, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2007-05-25 | | 公開日 | 2007-07-03 | | 最終更新日 | 2022-04-13 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal structure of a putative transcriptional regulator SCO0520 from Streptomyces coelicolor A3(2) reveals an unusual dimer among TetR family proteins.
J.Struct.Funct.Genom., 12, 2011
|
|
4WY2
 
 | | Crystal structure of universal stress protein E from Proteus mirabilis in complex with UDP-3-O-[(3R)-3-hydroxytetradecanoyl]-N-acetyl-alpha-glucosamine | | 分子名称: | CHLORIDE ION, GLYCEROL, SULFATE ION, ... | | 著者 | Shumilin, I.A, Shabalin, I.G, Handing, K.B, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2014-11-15 | | 公開日 | 2014-11-26 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal structure of universal stress protein E from Proteus mirabilis incomplex withUDP-3-O-[(3R)-3-hydroxytetradecanoyl]-N-acetyl-alpha-glucosamine
to be published
|
|
3QVM
 
 | | The structure of olei00960, a hydrolase from Oleispira antarctica | | 分子名称: | CALCIUM ION, CHLORIDE ION, Olei00960, ... | | 著者 | Singer, A.U, Kagan, O, Kim, Y, Edwards, A.M, Joachimiak, A, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2011-02-25 | | 公開日 | 2011-04-13 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (1.998 Å) | | 主引用文献 | Genome sequence and functional genomic analysis of the oil-degrading bacterium Oleispira antarctica.
Nat Commun, 4, 2013
|
|
3SVI
 
 | | Structure of the Pto-binding domain of HopPmaL generated by limited thermolysin digestion | | 分子名称: | CHLORIDE ION, SODIUM ION, SULFATE ION, ... | | 著者 | Singer, A.U, Stein, A, Xu, X, Cui, H, Joachimiak, A, Edwards, A.M, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2011-07-12 | | 公開日 | 2011-08-10 | | 最終更新日 | 2017-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structural Analysis of HopPmaL Reveals the Presence of a Second Adaptor Domain Common to the HopAB Family of Pseudomonas syringae Type III Effectors.
Biochemistry, 51, 2012
|
|
3TJY
 
 | | Structure of the Pto-binding domain of HopPmaL generated by limited chymotrypsin digestion | | 分子名称: | CHLORIDE ION, Effector protein hopAB3, SULFATE ION | | 著者 | Singer, A.U, Stein, A, Xu, X, Cui, H, Joachimiak, A, Edwards, A.M, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2011-08-25 | | 公開日 | 2011-09-14 | | 最終更新日 | 2013-01-09 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structural analysis of HopPmaL reveals the presence of a second adaptor domain common to the HopAB family of Pseudomonas syringae type III effectors.
Biochemistry, 51, 2012
|
|
3RT7
 
 | | Crystal structure of tm0922, a fusion of a domain of unknown function and ADP/ATP-dependent NAD(P)H-hydrate dehydratase from Thermotoga maritima in complex with ADP-glucose | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE-GLUCOSE, GLYCEROL, MAGNESIUM ION, ... | | 著者 | Shumilin, I.A, Cymborowski, M, Lesley, S.A, Minor, W. | | 登録日 | 2011-05-03 | | 公開日 | 2011-06-22 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Identification of unknown protein function using metabolite cocktail screening.
Structure, 20, 2012
|
|
3ROG
 
 | | Crystal Structure of Mouse Apolipoprotein A-I Binding Protein in Complex with Thymidine 3'-monophosphate | | 分子名称: | Apolipoprotein A-I-binding protein, SULFATE ION, THYMIDINE-3'-PHOSPHATE | | 著者 | Shumilin, I.A, Jha, K.N, Cymborowski, M, Herr, J.C, Minor, W. | | 登録日 | 2011-04-25 | | 公開日 | 2012-07-18 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | Identification of unknown protein function using metabolite cocktail screening.
Structure, 20, 2012
|
|
3RS8
 
 | | Crystal structure of tm0922, a fusion of a domain of unknown function and ADP/ATP-dependent NAD(P)H-hydrate dehydratase from Thermotoga maritima in complex with ADP-ribose | | 分子名称: | ADENOSINE-5-DIPHOSPHORIBOSE, GLYCEROL, MAGNESIUM ION, ... | | 著者 | Shumilin, I.A, Cymborowski, M, Lesley, S.A, Minor, W. | | 登録日 | 2011-05-02 | | 公開日 | 2011-06-22 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Identification of unknown protein function using metabolite cocktail screening.
Structure, 20, 2012
|
|
3RO7
 
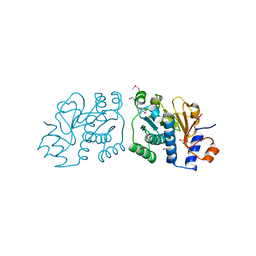 | | Crystal Structure of Mouse Apolipoprotein A-I Binding Protein in Complex with Thymine. | | 分子名称: | Apolipoprotein A-I-binding protein, SULFATE ION, THYMINE | | 著者 | Shumilin, I.A, Jha, K.N, Cymborowski, M, Herr, J.C, Minor, W. | | 登録日 | 2011-04-25 | | 公開日 | 2012-07-18 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Identification of unknown protein function using metabolite cocktail screening.
Structure, 20, 2012
|
|
3RSF
 
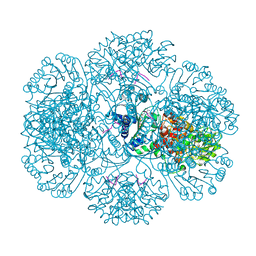 | | Crystal structure of tm0922, a fusion of a domain of unknown function and ADP/ATP-dependent NAD(P)H-hydrate dehydratase from Thermotoga maritima in complex with P1,P4-Di(adenosine-5') tetraphosphate | | 分子名称: | BIS(ADENOSINE)-5'-TETRAPHOSPHATE, POTASSIUM ION, Putative uncharacterized protein, ... | | 著者 | Shumilin, I.A, Cymborowski, M, Lesley, S.A, Minor, W. | | 登録日 | 2011-05-02 | | 公開日 | 2011-06-22 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Identification of unknown protein function using metabolite cocktail screening.
Structure, 20, 2012
|
|
3RTB
 
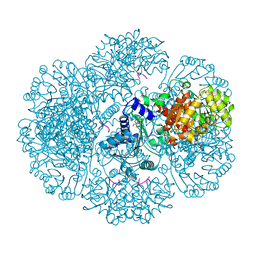 | | Crystal structure of tm0922, a fusion of a domain of unknown function and ADP/ATP-dependent NAD(P)H-hydrate dehydratase from Thermotoga maritima soaked with Adenosine-3'-5'-Diphosphate | | 分子名称: | ADENOSINE-3'-5'-DIPHOSPHATE, GLYCEROL, POTASSIUM ION, ... | | 著者 | Shumilin, I.A, Cymborowski, M, Lesley, S.A, Minor, W. | | 登録日 | 2011-05-03 | | 公開日 | 2011-07-27 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Identification of unknown protein function using metabolite cocktail screening.
Structure, 20, 2012
|
|
3RU2
 
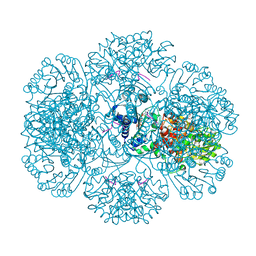 | | Crystal structure of tm0922, a fusion of a domain of unknown function and ADP/ATP-dependent NAD(P)H-hydrate dehydratase from Thermotoga maritima soaked with NADPH. | | 分子名称: | BETA-6-HYDROXY-1,4,5,6-TETRAHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE PHOSPHATE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, POTASSIUM ION, ... | | 著者 | Shumilin, I.A, Cymborowski, M, Lesley, S.A, Minor, W. | | 登録日 | 2011-05-04 | | 公開日 | 2011-07-27 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Identification of unknown protein function using metabolite cocktail screening.
Structure, 20, 2012
|
|
3RTA
 
 | | Crystal structure of tm0922, a fusion of a domain of unknown function and ADP/ATP-dependent NAD(P)H-hydrate dehydratase from Thermotoga maritima soaked with Acetyl Coenzyme A | | 分子名称: | ACETYL COENZYME *A, POTASSIUM ION, Putative uncharacterized protein, ... | | 著者 | Shumilin, I.A, Cymborowski, M, Lesley, S.A, Minor, W. | | 登録日 | 2011-05-03 | | 公開日 | 2011-06-22 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Identification of unknown protein function using metabolite cocktail screening.
Structure, 20, 2012
|
|
