8QBD
 
 | | Helical reconstruction of yeast eisosome protein Pil1 bound to membrane composed of lipid mixture +PIP2/+sterol (DOPC, DOPE, DOPS, cholesterol, PI(4,5)P2 35:20:20:15:10) | | 分子名称: | D-MYO-INOSITOL-1,4,5-TRIPHOSPHATE, O-[(R)-{[(2R)-2,3-bis(octadecanoyloxy)propyl]oxy}(hydroxy)phosphoryl]-L-serine, Sphingolipid long chain base-responsive protein PIL1 | | 著者 | Kefauver, J.M, Zou, L, Desfosses, A, Loewith, R.J. | | 登録日 | 2023-08-24 | | 公開日 | 2024-07-24 | | 最終更新日 | 2024-08-28 | | 実験手法 | ELECTRON MICROSCOPY (3.61 Å) | | 主引用文献 | Cryo-EM architecture of a near-native stretch-sensitive membrane microdomain.
Nature, 632, 2024
|
|
8QBF
 
 | | Compact state - Pil1 dimer with lipid headgroups fitted in native eisosome lattice bound to plasma membrane microdomain | | 分子名称: | D-MYO-INOSITOL-1,4,5-TRIPHOSPHATE, PHOSPHOSERINE, Sphingolipid long chain base-responsive protein PIL1 | | 著者 | Kefauver, J.M, Zou, L, Desfosses, A, Loewith, R.J. | | 登録日 | 2023-08-24 | | 公開日 | 2024-07-24 | | 最終更新日 | 2024-08-28 | | 実験手法 | ELECTRON MICROSCOPY (3.67 Å) | | 主引用文献 | Cryo-EM architecture of a near-native stretch-sensitive membrane microdomain.
Nature, 632, 2024
|
|
8QBG
 
 | |
8QBE
 
 | |
8QB8
 
 | |
6E5G
 
 | |
6EYC
 
 | | Re-refinement of the MCM2-7 double hexamer using ISOLDE | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, DNA replication licensing factor MCM2, DNA replication licensing factor MCM3, ... | | 著者 | Croll, T.I. | | 登録日 | 2017-11-11 | | 公開日 | 2018-06-20 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | ISOLDE: a physically realistic environment for model building into low-resolution electron-density maps.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
1QMN
 
 | |
481D
 
 | |
2XN6
 
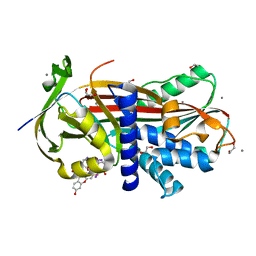 | | Crystal structure of thyroxine-binding globulin complexed with thyroxine-fluoresein | | 分子名称: | 1,2-ETHANEDIOL, 3',6'-DIHYDROXY-3-OXO-3H-SPIRO[2-BENZOFURAN-1,9'-XANTHENE]-6-CARBOXYLIC ACID, 3,5,3',5'-TETRAIODO-L-THYRONINE, ... | | 著者 | Qi, X, Yan, Y, Wei, Z, Zhou, A. | | 登録日 | 2010-07-30 | | 公開日 | 2011-02-16 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.29 Å) | | 主引用文献 | Allosteric Modulation of Hormone Release from Thyroxine and Corticosteroid Binding-Globulins.
J.Biol.Chem., 286, 2011
|
|
2XN7
 
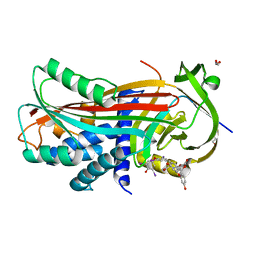 | | Crystal structure of thyroxine-binding globulin complexed with thyroxine-fluoresein (T405-CF) | | 分子名称: | 1,2-ETHANEDIOL, 3',6'-DIHYDROXY-3-OXO-3H-SPIRO[2-BENZOFURAN-1,9'-XANTHENE]-5-CARBOXYLIC ACID, 3,5,3',5'-TETRAIODO-L-THYRONINE, ... | | 著者 | Qi, X, Yan, Y, Wei, Z, Zhou, A. | | 登録日 | 2010-07-30 | | 公開日 | 2011-02-16 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.43 Å) | | 主引用文献 | Allosteric Modulation of Hormone Release from Thyroxine and Corticosteroid Binding-Globulins.
J.Biol.Chem., 286, 2011
|
|
2XN5
 
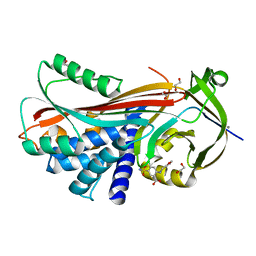 | | Crystal structure of thyroxine-binding globulin complexed with Furosemide | | 分子名称: | 1,2-ETHANEDIOL, 5-(AMINOSULFONYL)-4-CHLORO-2-[(2-FURYLMETHYL)AMINO]BENZOIC ACID, CALCIUM ION, ... | | 著者 | Qi, X, Yan, Y, Wei, Z, Zhou, A. | | 登録日 | 2010-07-30 | | 公開日 | 2011-02-16 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Allosteric Modulation of Hormone Release from Thyroxine and Corticosteroid Binding-Globulins.
J.Biol.Chem., 286, 2011
|
|
2XN3
 
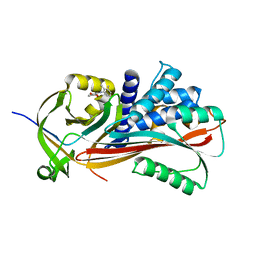 | | Crystal structure of thyroxine-binding globulin complexed with mefenamic acid | | 分子名称: | 2-[(2,3-DIMETHYLPHENYL)AMINO]BENZOIC ACID, THYROXINE-BINDING GLOBULIN | | 著者 | Qi, X, Yan, Y, Wei, Z, Zhou, A. | | 登録日 | 2010-07-30 | | 公開日 | 2011-02-16 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.09 Å) | | 主引用文献 | Allosteric Modulation of Hormone Release from Thyroxine and Corticosteroid Binding-Globulins.
J.Biol.Chem., 286, 2011
|
|
6P7M
 
 | | Cryo-EM structure of LbCas12a-crRNA: AcrVA4 (1:2 complex) | | 分子名称: | Cas12a, MAGNESIUM ION, anti-CRISPR VA4, ... | | 著者 | Knott, G.J, Liu, J.J, Doudna, J.A. | | 登録日 | 2019-06-06 | | 公開日 | 2019-08-21 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Structural basis for AcrVA4 inhibition of specific CRISPR-Cas12a.
Elife, 8, 2019
|
|
6P7N
 
 | | Cryo-EM structure of LbCas12a-crRNA: AcrVA4 (2:2 complex) | | 分子名称: | Cas12a, MAGNESIUM ION, anti-CRISPR VA4, ... | | 著者 | Knott, G.J, Liu, J.J, Doudna, J.A. | | 登録日 | 2019-06-06 | | 公開日 | 2019-08-21 | | 最終更新日 | 2023-08-16 | | 実験手法 | ELECTRON MICROSCOPY (4.9 Å) | | 主引用文献 | Structural basis for AcrVA4 inhibition of specific CRISPR-Cas12a.
Elife, 8, 2019
|
|
5V74
 
 | |
5TIS
 
 | | Room temperature XFEL structure of the native, doubly-illuminated photosystem II complex | | 分子名称: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | 著者 | Young, I.D, Ibrahim, M, Chatterjee, R, Gul, S, Fuller, F, Koroidov, S, Brewster, A.S, Tran, R, Alonso-Mori, R, Kroll, T, Michels-Clark, T, Laksmono, H, Sierra, R.G, Stan, C.A, Hussein, R, Zhang, M, Douthit, L, Kubin, M, de Lichtenberg, C, Pham, L.V, Nilsson, H, Cheah, M.H, Shevela, D, Saracini, C, Bean, M.A, Seuffert, I, Sokaras, D, Weng, T.-C, Pastor, E, Weninger, C, Fransson, T, Lassalle, L, Braeuer, P, Aller, P, Docker, P.T, Andi, B, Orville, A.M, Glownia, J.M, Nelson, S, Sikorski, M, Zhu, D, Hunter, M.S, Aquila, A, Koglin, J.E, Robinson, J, Liang, M, Boutet, S, Lyubimov, A.Y, Uervirojnangkoorn, M, Moriarty, N.W, Liebschner, D, Afonine, P.V, Watermann, D.G, Evans, G, Wernet, P, Dobbek, H, Weis, W.I, Brunger, A.T, Zwart, P.H, Adams, P.D, Zouni, A, Messinger, J, Bergmann, U, Sauter, N.K, Kern, J, Yachandra, V.K, Yano, J. | | 登録日 | 2016-10-03 | | 公開日 | 2016-11-23 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.25000381 Å) | | 主引用文献 | Structure of photosystem II and substrate binding at room temperature.
Nature, 540, 2016
|
|
6XR3
 
 | |
6XA9
 
 | | SARS CoV-2 PLpro in complex with ISG15 C-terminal domain propargylamide | | 分子名称: | GLYCEROL, ISG15 CTD-propargylamide, Non-structural protein 3, ... | | 著者 | Klemm, T, Calleja, D.J, Richardson, L.W, Lechtenberg, B.C, Komander, D. | | 登録日 | 2020-06-04 | | 公開日 | 2020-06-17 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Mechanism and inhibition of the papain-like protease, PLpro, of SARS-CoV-2.
Embo J., 39, 2020
|
|
5H31
 
 | |
5HLL
 
 | |
1EZX
 
 | |
8SP7
 
 | |
8TIR
 
 | | Isoreticular, interpenetrating co-crystal of Replication Initiator Protein REPE54 and symmetrical expanded duplex (31mer) containing the cognate REPE54 sequence and an additional G-C rich sequence with 1 sticky bases and 5' terminal phosphates. | | 分子名称: | DNA (5'-D(A*CP*CP*CP*GP*GP*AP*CP*CP*TP*GP*TP*GP*AP*CP*AP*AP*AP*TP*TP*GP*CP*CP*CP*TP*CP*AP*GP*AP*CP*GP*G)-3'), DNA (5'-D(A*GP*CP*CP*GP*TP*CP*TP*GP*AP*GP*GP*GP*CP*AP*AP*TP*TP*TP*GP*TP*CP*AP*CP*AP*GP*GP*TP*CP*CP*GP*G)-3'), MAGNESIUM ION, ... | | 著者 | Orun, A.R, Shields, E.T, Shrestha, R, Slaughter, C.K, Snow, C.D. | | 登録日 | 2023-07-20 | | 公開日 | 2023-08-02 | | 最終更新日 | 2024-08-14 | | 実験手法 | X-RAY DIFFRACTION (3.11 Å) | | 主引用文献 | Tuning chemical DNA ligation within DNA crystals and protein-DNA co-crystals
Acs Nanosci Au, 2024
|
|
8TIU
 
 | | Isoreticular, interpenetrating co-crystal of Replication Initiator Protein REPE54 and symmetrical expanded duplex (31mer) containing the cognate REPE54 sequence and an additional G-C rich sequence with 2 sticky base overhangs and 3' terminal phosphates. | | 分子名称: | DNA (5'-D(CP*CP*CP*GP*GP*AP*CP*CP*TP*GP*TP*GP*AP*CP*AP*AP*AP*TP*TP*GP*CP*CP*CP*TP*CP*AP*GP*AP*CP*GP*GP*A)-3')|, DNA (5'-D(GP*GP*CP*CP*GP*TP*CP*TP*GP*AP*GP*GP*GP*CP*AP*AP*TP*TP*TP*GP*TP*CP*AP*CP*AP*GP*GP*TP*CP*CP*GP*A)-3'), MAGNESIUM ION, ... | | 著者 | Orun, A.R, Shields, E.T, Shrestha, R, Slaughter, C.K, Snow, C.D. | | 登録日 | 2023-07-20 | | 公開日 | 2023-08-02 | | 最終更新日 | 2024-08-14 | | 実験手法 | X-RAY DIFFRACTION (3.9 Å) | | 主引用文献 | Tuning chemical DNA ligation within DNA crystals and protein-DNA co-crystals
Acs Nanosci Au, 2024
|
|
